molecule: intron 3 (i-ppo) encoded endonuclease;
synonym: intron-encoded endonuclease i-ppoi
|
|
| 3D3D | (3) | INTRON-ENCODED ENDONUCLEASE I-PPOI COMPLEXED WITH DNA |
|---|
| 3D3D | (2) | I-PPOL HOMING ENDONUCLEASE/DNA COMPLEX |
|---|
 | titl: dna binding and cleavage by the nuclear intron-encoded homing endonuclease i-ppoi. | |
| molecule: intron-encoded endonuclease i-ppoi; synonym: intron-encoded endonuclease i-ppoi |
| 3D3D | (1) | GROUP I MOBILE INTRON ENDONUCLEASE |
|---|
 | titl: the structure of i-crel, a group i intron-encoded homing endonuclease. |
| 3D3D | (1) | NOVOBIOCIN-RESISTANT MUTANT (R136H) OF THE N-TERMINAL 24 KDA FRAGMENT OF DNA GYRASE B COMPLEXED WITH NOVOBIOCIN AT 2.3 ANGSTROMS RESOLUTION |
|---|
| 3D3D | (1) | HUMAN URACIL-DNA GLYCOSYLASE |
|---|
| 3D3D | (2) | X-RAY STRUCTURE OF A POKEWEED ANTIVIRAL PROTEIN, CODED BY A NEW GENOMIC CLONE, AT 0.23 NM RESOLUTION. A MODEL STRUCTURE PROVIDES A SUITABLE ELECTROSTATIC FIELD FOR SUBSTRATE BINDING. |
|---|
| 3D3D | (1) | TRANSCRIPTIONAL REPRESSOR COPG/DNA COMPLEX |
|---|
 | titl: the structure of plasmid-encoded transcriptional repressor copg unliganded and bound to its operator. |
| 3D3D | (1) | STRUCTURAL COMPARISON OF AMYLOIDOGENIC LIGHT CHAIN DIMER IN TWO CRYSTAL FORMS WITH NONAMYLOIDOGENIC COUNTERPARTS |
|---|
 | expression_system_gene: cdna (genbank accession code u31344) |
| 3D3D | (4) | I-DMOI, INTRON-ENCODED ENDONUCLEASE |
|---|
| 3D3D | (1) | HTAFII18/HTAFII28 HETERODIMER CRYSTAL STRUCTURE |
|---|
 | titl: human taf(ii)28 and taf(ii)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the spt3 family. |
| 3D3D | (1) | HTAFII18/HTAFII28 HETERODIMER CRYSTAL STRUCTURE WITH BOUND PCMBS |
|---|
 | titl: human taf(ii)28 and taf(ii)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the spt3 family. |
| 3D3D | (1) | IMMUNOGLOBULIN LIGHT CHAIN PROTEIN |
|---|
 | expression_system_gene: cdna (genbank accession code u31344); gene: cdna (genbank accession code |
| 3D3D | (2) | PUTATIVE ANCESTRAL PROTEIN ENCODED BY A SINGLE SEQUENCE REPEAT OF THE MULTIDOMAIN PROTEINASE INHIBITOR FROM NICOTIANA ALATA |
|---|
| 3D3D | (2) | INTRON ENCODED HOMING ENDONUCLEASE I-PPOI (H98A)/DNA HOMING SITE COMPLEX |
|---|
 | intron encoded homing endonuclease i-ppoi (h98a)/dna homing site complex | |
| molecule: intron-encoded homing endonuclease i-ppoi |
| 3D3D | (2) | INTRON ENCODED HOMING ENDONUCLEASE I-PPOI/DNA COMPLEX LACKING CATALYTIC METAL ION |
|---|
 | intron encoded homing endonuclease i-ppoi/dna complex lacking catalytic metal ion | |
| molecule: intron-encoded homing endonuclease i-ppoi |
| 3D3D | (1) | CRYSTAL STRUCTURE AT 2.7A RESOLUTION OF A COMPLEX BETWEEN A STAPHYLOCOCCUS AUREUS DOMAIN AND A FAB FRAGMENT OF A HUMAN IGM ANTIBODY |
|---|
| 3D3D | (3) | CRYSTAL STRUCTURE OF AN ARCHAEAL INTEIN-ENCODED HOMING ENDONUCLEASE PI-PFUI |
|---|
| 3D3D | (1) | RUBREDOXIN FROM GUILLARDIA THETA |
|---|
 | other_details: the rbx-gene is encoded on the g.theta nucleomorph (chromosome ii) |
| 3D3D | (1) | THE 1.7 A CRYSTAL STRUCTURE OF A BLEOMYCIN RESISTANCE DETERMINANT ENCODED ON THE TRANSPOSON TN5 |
|---|
 | the 1.7 a crystal structure of a bleomycin resistance determinant encoded on the transposon tn5 |
| 3D3D | (1) | THE X-RAY CRYSTALLOGRAPHIC STRUCTURE OF BETA CARBONIC ANHYDRASE FROM THE C3 DICOT PISUM SATIVUM |
|---|
 | cellular_location: nuclear encoded |
| 3D3D | (1) | CRYSTAL STRUCTURE OF SALMONELLA TYPHIMURIUM CU,ZN SUPEROXIDE DISMUTASE |
|---|
 | titl: functional and crystallographic characterization of salmonella typhimurium cu,zn superoxide dismutase coded by the sodci virulence gene. |
| 3D3D | (1) | L116A MUTANT OF THE HOMING ENDONUCLEASE I-PPOI COMPLEXED TO HOMING SITE DNA. |
|---|
 | synonym: i-ppoi, intron-encoded endonuclease i-ppoi |
| 3D3D | (1) | APO CRYSTAL STRUCTURE OF THE HOMING ENDONUCLEASE, I-PPOI |
|---|
 | molecule: intron-encoded homing endonuclease i-ppoi |
| 3D3D | (2) | STRUCTURE OF ESCHERICHIA COLI PURT-ENCODED GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE COMPLEXED WITH MG AND AMPPNP |
|---|
| 3D3D | (2) | STRUCTURE OF ESCHERICHIA COLI PURT-ENCODED GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE COMPLEXED WITH MG, AMPPNP, AND GAR |
|---|
| 3D3D | (1) | SOLUTION STRUCTURE OF FKBP12 COMPLEXED WITH GPI-1046, A NEUROTROPHIC LIGAND |
|---|
 | other_details: protein coordinates were taken from the crystal structure by holt et al. (1993, pdb accession code 1fkg). |
| 3D3D | (1) | FHUA IN COMPLEX WITH LIPOPOLYSACCHARIDE AND RIFAMYCIN CGP4832 |
|---|
 | expression_system_vector: plasmid-encoded hexahistidine-tagged fhua |
| 3D3D | (1) | STRUCTURE OF A CYS2HIS2 ZINC FINGER/TATA BOX COMPLEX (CLONE #2) |
|---|
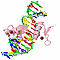 | titl: beyond the "recognition code": structures of two cys2his2 zinc finger/tata box complexes. |
| 3D3D | (1) | STRUCTURE OF A CYS2HIS2 ZINC FINGER/TATA BOX COMPLEX (TATAZF;CLONE #6) |
|---|
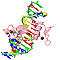 | titl: beyond the "recognition code": structures of two cys2his2 zinc finger/tata box complexes. |
| 3D3D | (1) | STRUCTURE OF CYTOSOLIC PROTEIN OF UNKNOWN FUNCTION CODED BY GENE FROM NUSA/INFB REGION, A YLXR HOMOLOGUE |
|---|
 | structure of cytosolic protein of unknown function coded by gene from nusa/infb region, a ylxr homologue |
| 3D3D | (1) | LAGLIDADG HOMING ENDONUCLEASE I-CREI / DNA PRODUCT COMPLEX WITH MAGNESIUM |
|---|
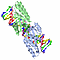 | gene: encoded by an orf within cr.lsu intron of chloroplast 23s rrna gene |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE PLASMID MAINTENANCE SYSTEM EPSILON/ZETA: MEACHNISM OF TOXIN INACTIVATION AND TOXIN FUNCTION |
|---|
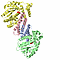 | other_details: encoded on plasmid psm19035 |
| 3D3D | (1) | THREE-DIMENSIONAL STRUCTURE OF THE QUORUM SENSING PROTEIN TRAR BOUND TO ITS AUTOINDUCER AND TO ITS TARGET DNA |
|---|
 | other_details: encoded on plasmid ptia6nc |
| 3D3D | (2) | THEORETICAL MODEL OF THE BETA-HELICAL DOMAIN OF STREPTOMYCES SP. MYCODEXTRANASE |
|---|
 | theoretical model of the beta-helical domain of streptomyces sp. mycodextranase | |
| molecule: mycodextranase |
| 3D3D | (1) | RUBREDOXIN FROM GUILLARDIA THETA |
|---|
 | other_details: the rbx-gene is encoded on the g.theta nucleomorph (chromosome ii) |
| 3D3D | (2) | SOLUTION STRUCTURE OF HUMAN P8-MTCP1, A CYSTEINE-RICH PROTEIN ENCODED BY THE MTCP1 ONCOGENE,REVEALS A NEW ALPHA-HELICAL ASSEMBLY MOTIF, NMR, MINIMIZED AVERAGE STRUCTURE |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF HLA-A*0201/MAGE-A4-PEPTIDE COMPLEX |
|---|
 | titl: high-resolution structure of hla-a*0201 in complex with a tumour-specific antigenic peptide encoded by the mage-a4 gene. |
| 3D3D | (2) | SOLUTION STRUCTURE AND FUNCTION OF A CONSERVED PROTEIN SP14.3 ENCODED BY AN ESSENTIAL STREPTOCOCCUS PNEUMONIAE GENE |
|---|
| 3D3D | (1) | C-TERMINAL DOMAIN OF TRANSCRIPTIONAL REPRESSOR PROTEIN KORB |
|---|
 | titl: an src homology 3-like domain is responsible for dimerization of the repressor protein korb encoded by the promiscuous incp plasmid rp4. |
| 3D3D | (1) | C-TERMINAL DOMAIN OF THE TRANSCRIPTIONAL REPRESSOR PROTEIN KORB |
|---|
 | titl: an src homology 3-like domain is responsible for dimerization of the repressor protein korb encoded by the promiscuous incp plasmid rp4. |
| 3D3D | (1) | GLYTM1BZIP: A CHIMERIC PEPTIDE MODEL OF THE N-TERMINUS OF A RAT SHORT ALPHA TROPOMYOSIN WITH THE N-TERMINUS ENCODED BY EXON 1B |
|---|
 | glytm1bzip: a chimeric peptide model of the n-terminus of a rat short alpha tropomyosin with the n-terminus encoded by exon 1b |
| 3D3D | (2) | HOMING ENDONUCLEASE/DNA COMPLEX |
|---|
 | titl: dna binding and cleavage by the nuclear intron-encoded homing endonuclease i-ppoi. | |
| molecule: intron-encoded endonuclease i-ppoi |
| 3D3D | (1) | CRYSTAL STRUCTURE OF OMEGA TRANSCRIPTIONAL REPRESSOR AT 1.5A RESOLUTION |
|---|
 | titl: crystal structure of omega transcriptional repressor encoded by streptococcus pyogenes plasmid psm19035 at 1.5 a resolution. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF ARCHAEAL TYROSYL-TRNA SYNTHETASE COMPLEXED WITH TRNA(TYR) AND L-TYROSINE |
|---|
 | titl: structural basis for orthogonal trna specificities of tyrosyl-trna synthetases for genetic code expansion |
| 3D3D | (2) | CRYSTAL STRUCTURE OF GP2.5, A SINGLE-STRANDED DNA BINDING PROTEIN ENCODED BY BACTERIOPHAGE T7 |
|---|
| 3D3D | (1) | STRUCTURE OF PUTATIVE ASPARAGINASE ENCODED BY ESCHERICHIA COLI YBIK GENE |
|---|
 | structure of putative asparaginase encoded by escherichia coli ybik gene |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A COMPLEX OF HUMAN CDK6 AND A VIRAL CYCLIN |
|---|
 | titl: structural basis for cdk6 activation by a virus-encoded cyclin. |
| 3D3D | (1) | ARE CARBOXY TERMINII OF HELICES CODED BY THE LOCAL SEQUENCE OR BY TERTIARY STRUCTURE CONTACTS |
|---|
 | are carboxy terminii of helices coded by the local sequence or by tertiary structure contacts |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE ANKYRIN REPEAT DOMAIN OF BCL-3: A UNIQUE MEMBER OF THE IKAPPAB PROTEIN FAMILY |
|---|
 | molecule: b-cell lymphoma 3-encoded protein |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE ANKYRIN REPEAT DOMAIN OF BCL-3: A UNIQUE MEMBER OF THE IKAPPAB PROTEIN FAMILY |
|---|
 | molecule: b-cell lymphoma 3-encoded protein |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PUTATIVE ASPARAGINASE ENCODED BY ESCHERICHIA COLI YBIK GENE |
|---|
 | crystal structure of putative asparaginase encoded by escherichia coli ybik gene |
| 3D3D | (1) | BACILLUS CIRCULANS STRAIN 251 CYCLODEXTRIN GLYCOSYL TRANSFERASE MUTANT N193G |
|---|
 | glycosyl transferase, transferase, cylcodextrin, acarbose |
| 3D3D | (2) | CRYSTAL STRUCTURE OF PURT-ENCODED GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE IN COMPLEX WITH MG-ATP AND GAR |
|---|
| 3D3D | (2) | CRYSTAL STRUCTURE OF PURT-ENCODED GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE COMPLEXED WITH MG-ATP |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE IN COMPLEX WITH MG-AMPPCP |
|---|
 | titl: purt-encoded glycinamide ribonucleotide transformylase. accommodation of adenosine nucleotide analogs within the active site. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF GLYCNIAMIDE RIBONUCLEOTIDE TRANSFORMYLASE IN COMPLEX WITH MG-ATP-GAMMA-S |
|---|
 | titl: purt-encoded glycinamide ribonucleotide transformylase. accommodation of adenosine nucleotide analogs within the active site. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE IN COMPLEX WITH MG-ADP |
|---|
 | titl: purt-encoded glycinamide ribonucleotide transformylase. accommodation of adenosine nucleotide analogs within the active site. |
| 3D3D | (3) | STRUCTURE AND FUNCTION OF A VIRALLY ENCODED FUNGAL TOXIN FROM USTILAGO MAYDIS: A FUNGAL AND MAMMALIAN CALCIUM CHANNEL INHIBITOR |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE METHANOSARCINA BARKERI MONOMETHYLAMINE METHYLTRANSFERASE (MTMB) |
|---|
 | titl: a new uag-encoded residue in the structure of a methanogen methyltransferase. |
| 3D3D | (1) | NON STRUCTURAL PROTEIN ENCODED BY GENE SEGMENT 8 OF ROTAVIRUS (NSP2), AN NTPASE, SSRNA BINDING AND NUCLEIC ACID HELIX-DESTABILIZING PROTEIN |
|---|
 | non structural protein encoded by gene segment 8 of rotavirus (nsp2), an ntpase, ssrna binding and nucleic acid helix-destabilizing protein |
| 3D3D | (1) | ARE CARBOXY TERMINII OF HELICES CODED BY THE LOCAL SEQUENCE OR BY TERTIARY STRUCTURE CONTACTS |
|---|
 | are carboxy terminii of helices coded by the local sequence or by tertiary structure contacts |
| 3D3D | (1) | HIGH RESOLUTION STRUCTURE OF FOSFOMYCIN RESISTANCE PROTEIN A (FOSA) |
|---|
 | titl: crystal structure of a genomically encoded fosfomycin resistance protein (fosa) at 1.19 a resolution by mad phasing off the l-iii edge of tl(+) |
| 3D3D | (1) | CRYSTAL STRUTCURE OF THE FOSFOMYCIN RESISTANCE PROTEIN A (FOSA) CONTAINING BOUND THALLIUM CATIONS |
|---|
 | titl: crystal structure of a genomically encoded fosfomycin resistance protein (fosa) at 1.19 a resolution by mad phasing off the l-iii edge of tl(+) |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE FOSFOMYCIN RESISTANCE PROTEIN (FOSA) CONTAINING BOUND SUBSTRATE |
|---|
 | titl: crystal structure of a genomically encoded fosfomycin resistance protein (fosa) at 1.19 a resolution by mad phasing off the l-iii edge of tl(+) |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A BACTERIAL GLUTATHIONE TRANSFERASE FROM ESCHERICHIA COLI WITH GLUTATHIONE SULFONATE IN THE ACTIVE SITE |
|---|
 | titl: conserved structural elements in glutathione transferase homologues encoded in the genome of escherichia coli |
| 3D3D | (1) | SOLUTION NMR STRUCTURE OF PROTEIN YOAG FROM ESCHERICHIA COLI. ONTARIO CENTRE FOR STRUCTURAL PROTEOMICS TARGET EC0264_1_60; NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET94. |
|---|
 | titl: solution structure of hypothetical protein dimer encoded by the yoag gene from escherichia coli |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE METHANOSARCINA BARKERI MONOMETHYLAMINE METHYLTRANSFERASE (MTMB) |
|---|
 | titl: a new uag-encoded residue in the structure of a methanogen methyltransferase |
| 3D3D | (1) | SOLUTION NMR STRUCTURE OF PROTEIN AQ_1857 FROM AQUIFEX AEOLICUS: NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET QR6. |
|---|
 | titl: nmr structure of the hypothetical protein aq-1857 encoded by the y157 gene from aquifex aeolicus reveals a novel protein fold. |
| 3D3D | (1) | COMPLEX BETWEEN BAFF AND A BR3 DERIVED PEPTIDE PRESENTED IN A BETA-HAIRPIN SCAFFOLD |
|---|
 | titl: baff/blys receptor 3 comprises a minimal tnf receptor-like module that encodes a highly focused ligand-binding site |
| 3D3D | (1) | SOLUTION STRUCTURE OF THE EXTRACELLULAR DOMAIN OF BLYS RECEPTOR 3 (BR3) |
|---|
 | titl: baff/blys receptor 3 comprises a minimal tnf receptor-like module that encodes a highly focused ligand-binding site |
| 3D3D | (3) | EXPANSION OF THE GENETIC CODE ENABLES DESIGN OF A NOVEL "GOLD" CLASS OF GREEN FLUORESCENT PROTEINS |
|---|
| 3D3D | (3) | EXPANSION OF THE GENETIC CODE ENABLES DESIGN OF A NOVEL "GOLD" CLASS OF GREEN FLUORESCENT PROTEINS |
|---|
| 3D3D | (3) | EXPANSION OF THE GENETIC CODE ENABLES DESIGN OF A NOVEL "GOLD" CLASS OF GREEN FLUORESCENT PROTEINS |
|---|
| 3D3D | (1) | THE STRUCTURE AND DNA RECOGNITION OF A BIFUNCTIONAL HOMING ENDONUCLEASE AND GROUP I INTRON SPLICING FACTOR |
|---|
 | molecule: intron-encoded endonuclease i-anii |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A CU-ZN SUPEROXIDE DISMUTASE FROM MYCOBACTERIUM TUBERCULOSIS AT 1.63 RESOLUTION |
|---|
 | titl: unique features of the sodc-encoded superoxide dismutase from mycobacterium tuberculosis, a fully functional copper-containing enzyme lacking zinc in the active site. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF AN ACTIVE FRAGMENT OF HUMAN TYROSYL-TRNA SYNTHETASE WITH TYROSINOL |
|---|
 | titl: crystal structures that suggest late development of genetic code components for differentiating aromatic side chains |
| 3D3D | (1) | THE CRYSTAL STRUCTURE OF FIBROBLAST GROWTH FACTOR 7/1 CHIMERA |
|---|
 | fragment: dual-function chimera between rat fgf-7 encoded by exon 1 and 2 and human fgf-1 encoded by exon 3 |
| 3D3D | (1) | SOLUTION STRUCTURE OF 30S RIBOSOMAL PROTEIN S27E FROM ARCHAEOGLOBUS FULGIDUS: GR2, A NESG TARGET PROTEIN |
|---|
 | titl: the nmr solution structure of the 30s ribosomal protein s27e encoded in gene rs27_arcfu of archaeoglobus fulgidis reveals a novel protein fold |
| 3D3D | (1) | STRUCTURE OF YCFC PROTEIN OF UNKNOWN FUNCTION ESCHERICHIA COLI |
|---|
 | titl: structure of a hypothetical protein ycfc coded by escherichia coli genome. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HUMAN TRYPTOPHANYL-TRNA SYNTHETASE |
|---|
 | titl: crystal structures that suggest late development of genetic code components for differentiating aromatic side chains |
| 3D3D | (1) | THE HOMING ENDONUCLEASE I-SCEI BOUND TO ITS DNA RECOGNITION REGION |
|---|
 | molecule: intron-encoded endonuclease i-scei |
| 3D3D | (1) | CRYSTAL STRUCTURE OF TRAC |
|---|
 | titl: structural and functional characterization of the virb5 protein from the type iv secretion system encoded by the conjugative plasmid pkm101 |
| 3D3D | (2) | PROBING THE ROLE OF TRYPTOPHANS IN AEQUOREA VICTORIA GREEN FLUORESCENT PROTEINS WITH AN EXPANDED GENETIC CODE |
|---|
| 3D3D | (2) | PROBING THE ROLE OF TRYPTOPHANS IN AEQUOREA VICTORIA GREEN FLUORESCENT PROTEINS WITH AN EXPANDED GENETIC CODE |
|---|
| 3D3D | (2) | PROBING THE ROLE OF TRYPTOPHANS IN AEQUOREA VICTORIA GREEN FLUORESCENT PROTEINS WITH AN EXPANDED GENETIC CODE |
|---|
| 3D3D | (2) | PROBING THE ROLE OF TRYPTOPHANS IN AEQUOREA VICTORIA GREEN FLUORESCENT PROTEINS WITH AN EXPANDED GENETIC CODE |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF CAG-Z FROM HELICOBACTER PYLORI |
|---|
 | titl: crystal structure of cagz, a protein from the helicobacter pylori pathogenicity island that encodes for a type iv secretion system |
| 3D3D | (2) | SOLUTION STRUCTURE OF THE E. COLI BACTERIOPHAGE P1 ENCODED HOT PROTEIN: A HOMOLOGUE OF THE THETA SUBUNIT OF E. COLI DNA POLYMERASE III |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE DNA-BINDING DOMAIN OF INTRON ENDONUCLEASE I-TEVI WITH OPERATOR SITE |
|---|
 | titl: intron-encoded homing endonuclease i-tevi also functions as a transcriptional autorepressor. |
| 3D3D | (1) | ALPHA-SPECTRIN SRC HOMOLOGY 3 DOMAIN, CIRCULAR PERMUTANT, CUT AT N47-D48 |
|---|
| 3D3D | (1) | STRUCTURE OF BAL32A FROM A SOIL-DERIVED MOBILE GENE CASSETTE |
|---|
 | titl: integron-associated mobile gene cassettes code for folded proteins: the structure of bal32a, a new member of the adaptable alpha+beta barrel family |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE HYDROXYLAMINE MTMB COMPLEX |
|---|
 | titl: reactivity and chemical synthesis of l-pyrrolysine- the 22(nd) genetically encoded amino acid |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE N-METHYL-HYDROXYLAMINE MTMB COMPLEX |
|---|
 | titl: reactivity and chemical synthesis of l-pyrrolysine- the 22(nd) genetically encoded amino acid |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE SULFITE MTMB COMPLEX |
|---|
 | titl: reactivity and chemical synthesis of l-pyrrolysine- the 22(nd) genetically encoded amino acid |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE FAB YADS1 COMPLEXED WITH H-VEGF |
|---|
 | titl: synthetic antibodies from a four-amino-acid code: a dominan role for tyrosine in antigen recognition |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE FAB YADS2 COMPLEXED WITH H-VEGF |
|---|
 | titl: synthetic antibodies from a four-amino-acid code: a dominan role for tyrosine in antigen recognition |
| 3D3D | (1) | THE STRUCTURAL BASIS OF SPECIFIC BASE EXCISION REPAIR BY URACIL-DNA GLYCOSYLASE |
|---|
 | other_details: encoded by the ul2 orf of herpes simplex virus type 1 |
| 3D3D | (1) | THE STRUCTURAL BASIS OF SPECIFIC BASE EXCISION REPAIR BY URACIL-DNA GLYCOSYLASE |
|---|
 | other_details: encoded by the ul2 orf of herpes simplex virus type 1 |
| 3D3D | (3) | CRYSTAL STRUCTURE OF GLUCODEXTRANASE FROM ARTHROBACTER GLOBIFORMIS I42 |
|---|
| 3D3D | (3) | CRYSTAL STRUCTURE OF GLUCODEXTRANASE COMPLEXED WITH ACARBOSE |
|---|
| 3D3D | (1) | COMPLEX OF ECHOVIRUS TYPE 12 WITH DOMAINS 3 AND 4 OF ITS RECEPTOR DECAY ACCELERATING FACTOR (CD55) BY CRYO ELECTRON MICROSCOPY AT 16 A |
|---|
 | other_details: structure of echovirus type 11 fitted into cryo-em electron density for echovirus type 12. the em density has been deposited in the emdb, with accession code 1057 |
| 3D3D | (1) | SPINACH RUBISCO IN COMPLEX WITH 2-CARBOXYARABINITOL 2 BISPHOSPHATE AND CALCIUM. |
|---|
| 3D3D | (1) | ESCHERICHIA COLI TYROSYL-TRNA SYNTHETASE MUTANT COMPLEXED WITH TYR-AMS |
|---|
 | titl: structural basis of nonnatural amino acid recognition by an engineered aminoacyl-trna synthetase for genetic code expansion |
| 3D3D | (1) | STRUCTURE OF DIHYDROFOLATE REDUCTASE |
|---|
 | titl: a plasmid-encoded dihydrofolate reductase from trimethoprim-resistant bacteria has a novel d2-symmetric active site. |
| 3D3D | (1) | STRUCTURE OF DIHYDROFOLATE REDUCTASE |
|---|
 | titl: a plasmid-encoded dihydrofolate reductase from trimethoprim-resistant bacteria has a novel d2-symmetric active site. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF ARGININE BIOSYNTHESIS BIFUNCTIONAL PROTEIN ARGJ (10175521) FROM BACILLUS HALODURANS AT 2.00 A RESOLUTION |
|---|
 | other_details: both chain a and chain b were expressed from a single construct which encodes residues 1-411 of the argj gene |
| 3D3D | (2) | IDENTIFICATION AND CHARACTERIZATION OF THE FIRST PLANT G-QUADRUPLEX BINDING PROTEIN ENCODED BY THE ZEA MAYS L. NUCLEOSIDE DIPHOSPHATE1 GENE, ZMNDPK1 |
|---|
| 3D3D | (1) | ENDONUCLEASE DOMAIN OF TRAS1, A TELOMERE-SPECIFIC NON-LTR RETROTRANSPOSON |
|---|
 | titl: crystal structure of the endonuclease domain encoded by the telomere-specific long interspersed nuclear element, tras1 |
| 3D3D | (1) | NMR STRUCTURE OF VONTR |
|---|
 | titl: conserved structural and sequence elements implicated in the processing of gene-encoded circular proteins |
| 3D3D | (1) | NMR STRUCTURE OF OANTR |
|---|
 | titl: conserved structural and sequence elements implicated in the processing of gene-encoded circular proteins |
| 3D3D | (1) | ESCHERICHIA COLI TYROSYL-TRNA SYNTHETASE MUTANT COMPLEXED WITH 3-IODO-L-TYROSINE |
|---|
 | titl: structural basis of nonnatural amino acid recognition by an engineered aminoacyl-trna synthetase for genetic code expansion |
| 3D3D | (1) | ESCHERICHIA COLI TYROSYL-TRNA SYNTHETASE MUTANT COMPLEXED WITH L-TYROSINE |
|---|
 | titl: structural basis of nonnatural amino acid recognition by an engineered aminoacyl-trna synthetase for genetic code expansion |
| 3D3D | (2) | TRANSCRIPTION FACTOR 1, NMR, MINIMIZED AVERAGE STRUCTURE |
|---|
| 3D3D | (1) | SOLUTION STRUCTURE OF NORTHEAST STRUCTURAL GENOMICS TARGET PROTEIN BCR68 ENCODED IN GENE Q816V6 OF B. CEREUS |
|---|
 | solution structure of northeast structural genomics target protein bcr68 encoded in gene q816v6 of b. cereus |
| 3D3D | (1) | STRUCTURE OF A HEMOLYSIN-COREGULATED PROTEIN FROM PSEUDOMONAS AERUGINOSA |
|---|
 | titl: a virulence locus of pseudomonas aeruginosa encodes a protein secretion apparatus. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF AZITHROMYCIN BOUND TO THE G2099A MUTANT 50S RIBOSOMAL SUBUNIT OF HALOARCULA MARISMORTUI |
|---|
 | other_details: encoded by rrna operon |
| 3D3D | (1) | CRYSTAL STRUCTURE OF ERYTHROMYCIN BOUND TO THE G2099A MUTANT 50S RIBOSOMAL SUBUNIT OF HALOARCULA MARISMORTUI |
|---|
 | other_details: encoded by rrna operon |
| 3D3D | (1) | CRYSTAL STRUCTURE OF TELITHROMYCIN BOUND TO THE G2099A MUTANT 50S RIBOSOMAL SUBUNIT OF HALOARCULA MARISMORTUI |
|---|
 | other_details: encoded by rrna operon |
| 3D3D | (1) | CRYSTAL STRUCTURE OF VIRGINIAMYCIN M AND S BOUND TO THE 50S RIBOSOMAL SUBUNIT OF HALOARCULA MARISMORTUI |
|---|
 | other_details: encoded by rrna operon |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE MUTANT 50S RIBOSOMAL SUBUNIT OF HALOARCULA MARISMORTUI CONTAINING A THREE RESIDUE DELETION IN L22 |
|---|
 | other_details: encoded by rrna operon |
| 3D3D | (1) | CRYSTAL STRUCTURE OF CLINDAMYCIN BOUND TO THE G2099A MUTANT 50S RIBOSOMAL SUBUNIT OF HALOARCULA MARISMORTUI |
|---|
 | other_details: encoded by rrna operon |
| 3D3D | (1) | CRYSTAL STRUCTURE OF QUINUPRISTIN BOUND TO THE G2099A MUTANT 50S RIBOSOMAL SUBUNIT OF HALOARCULA MARISMORTUI |
|---|
 | other_details: encoded by rrna operon |
| 3D3D | (2) | SOLUTION STRUCTURE OF THE SARS CORONAVIRUS ORF 7A CODED X4 PROTEIN |
|---|
| 3D3D | (1) | THE CRYSTAL STRUCTURE OF THE YSD1 FAB BOUND TO DR5 |
|---|
 | titl: molecular recognition by a binary code. |
| 3D3D | (2) | CRYSTAL STRUCTURE OF PLASMID-ENCODED CLASS C BETA-LACTAMASE CMY-2 COMPLEXED WITH CITRATE MOLECULE |
|---|
| 3D3D | (2) | STRUCTURAL BASIS FOR THE EXTENDED SUBSTRATE SPECTRUM OF CMY-10, A PLASMID-ENCODED CLASS C BETA-LACTAMASE |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF A VIRAL CHEMOKINE |
|---|
 | titl: crystal structure of viral macrophage inflammatory protein encoded by kaposi's sarcoma-associated herpesvirus at 1.7a. |
| 3D3D | (1) | SOLUTION STRUCTURE OF 50S RIBOSOMAL PROTEIN L40E FROM SULFOLOBUS SOLFATARICUS |
|---|
 | titl: solution structure of ribosomal protein l40e, a unique c4 zinc finger protein encoded by archaeon sulfolobus solfataricus |
| 3D3D | (1) | XPF FROM AEROPYRUM PERNIX, COMPLEX WITH DNA |
|---|
 | other_details: a pernix genome sequencing project gene ape1436, accession code c72622 |
| 3D3D | (1) | XPF FROM AEROPYRUM PERNIX |
|---|
 | other_details: a pernix genome sequencing project gne ape 1436, accession code c72622 |
| 3D3D | (2) | THEORETICAL INTERACTION MODEL OF PLASMEPSIN II FROM PLASMODIUM FALCIPARUM WITH A LIGAND FROM AN ECODED STATIN COMBINATORIAL LIBRARY |
|---|
| 3D3D | (1) | STRUCTURAL BASIS FOR COOPERATIVE BINDING OF RIBBON-HELIX-HELIX OMEGA REPRESSOR TO DIRECT DNA HEPTAD REPEATS |
|---|
 | other_details: omega transcriptional repressor is encoded by plasmid psm19035 of the inc18 family of plasmids |
| 3D3D | (1) | STRUCTURAL BASIS FOR COOPERATIVE BINDING OF RIBBON-HELIX-HELIX OMEGA REPRESSOR TO INVERTED DNA HEPTAD REPEATS |
|---|
 | other_details: omega transcriptional repressor is encoded by plasmid psm19035 of the inc18 family of plasmids |
| 3D3D | (2) | THE STRUCTURE OF A GROUP A STREPTOCOCCAL PHAGE-ENCODED TAIL-FIBRE SHOWING HYALURONAN LYASE ACTIVITY. |
|---|
| 3D3D | (1) | COMPLEX OF ECHOVIRUS TYPE 12 WITH DOMAINS 1, 2, 3 AND 4 OF ITS RECEPTOR DECAY ACCELERATING FACTOR (CD55) BY CRYO ELECTRON MICROSCOPY AT 16 A |
|---|
 | other_details: structure of echovirus type 11 fitted into cryo-em electron density for echovirus type 12. the em density has been deposited in the emdb, with accession code 1057 |
| 3D3D | (1) | STRUCTURAL BASIS FOR COOPERATIVE BINDING OF RIBBON-HELIX-HELIX REPRESSOR OMEGA TO MUTATED DIRECT DNA HEPTAD REPEATS |
|---|
 | other_details: omega transcriptional reprsssor is encoded by plasmid psm19035 of the inc18 family of plasmids. |
| 3D3D | (2) | STRUCTURE-BASED FUNCTIONAL ANNOTATION: YEAST YMR099C CODES FOR A D-HEXOSE-6-PHOSPHATE MUTAROTASE. |
|---|
| 3D3D | (2) | STRUCTURE-BASED FUNCTIONAL ANNOTATION: YEAST YMR099C CODES FOR A D-HEXOSE-6-PHOSPHATE MUTAROTASE. COMPLEX WITH GLUCOSE-6-PHOSPHATE |
|---|
| 3D3D | (2) | STRUCTURE-BASED FUNCTIONAL ANNOTATION: YEAST YMR099C CODES FOR A D-HEXOSE-6-PHOSPHATE MUTAROTASE. COMPLEX WITH TAGATOSE-6-PHOSPHATE |
|---|
| 3D3D | (1) | TRANSCRIPTIONAL REPRESSOR COPG |
|---|
 | titl: the structure of plasmid-encoded transcriptional repressor copg unliganded and bound to its operator. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF INTEIN HOMING ENDONUCLEASE II |
|---|
 | titl: crystal structure of intein homing endonuclease ii encoded in dna polymerase gene from hyperthermophilic archaeon thermococcus kodakaraensis strain kod1 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF INTEIN HOMING ENDONUCLEASE II |
|---|
 | titl: crystal structure of intein homing endonuclease ii encoded in dna polymerase gene from hyperthermophilic archaeon thermococcus kodakaraensis strain kod1 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF GLU-TRNA(GLN) AMIDOTRANSFERASE IN THE COMPLEX WITH TRNA(GLN) |
|---|
 | titl: structural basis of rna-dependent recruitment of glutamine to the genetic code |
| 3D3D | (2) | CRYSTAL STRUCTURE OF ARCHAEAL INTRON-ENCODED HOMING ENDONUCLEASE I-TSP061I |
|---|
 | crystal structure of archaeal intron-encoded homing endonuclease i-tsp061i | |
| synonym: archaeal intron-encoded dna endonuclease |
| 3D3D | (1) | SOLUTION STRUCTURE OF THE UBIQUITIN-ASSOCIATED DOMAIN OF HUMAN BMSC-UBP AND ITS COMPLEX WITH UBIQUITIN |
|---|
| 3D3D | (1) | XYLANASE II FROM TRICODERMA REESEI AT 100K |
|---|
 | xylanase ii from tricoderma reesei at 100k |
| 3D3D | (1) | XYLANASE II FROM TRICODERMA REESEI AT 293K |
|---|
 | xylanase ii from tricoderma reesei at 293k |
| 3D3D | (1) | CRYSTAL STRUCTURE OF FFRP-DM1 |
|---|
 | titl: a structural code for discriminating between transcription signals revealed by the feast/famine regulatory protein dm1 in complex with ligands |
| 3D3D | (1) | GROUP I INTRON-ENCODED HOMING ENDONUCLEASE I-CEUI COMPLEXED WITH DNA |
|---|
 | group i intron-encoded homing endonuclease i-ceui complexed with dna |
| 3D3D | (2) | SOLUTION STRUCTURE FOR THE PROTEIN CODED BY GENE LOCUS BB0938 OF BORDETELLA BRONCHISEPTICA. NORTHEAST STRUCTURAL GENOMICS TARGET BOR11. |
|---|
| 3D3D | (3) | SOLUTION STRUCTURE OF THE COMPLEX BETWEEN POXVIRUS-ENCODED CC CHEMOKINE INHIBITOR VCCI AND HUMAN MIP-1BETA, MINIMIZED AVERAGE STRUCTURE |
|---|
| 3D3D | (3) | SOLUTION STRUCTURE OF THE COMPLEX BETWEEN POXVIRUS-ENCODED CC CHEMOKINE INHIBITOR VCCI AND HUMAN MIP-1BETA, ENSEMBLE STRUCTURE |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF RNASE Z/TRNA(THR) COMPLEX |
|---|
| 3D3D | (1) | PNRA FROM TREPONEMA PALLIDUM AS PURIFIED FROM E. COLI (BOUND TO INOSINE) |
|---|
 | titl: the pnra (tp0319; tmpc) lipoprotein represents a new family of bacterial purine nucleoside receptor encoded within an atp-binding cassette (abc)-like operon in treponema pallidum |
| 3D3D | (1) | PNRA FROM TREPONEMA PALLIDUM COMPLEXED WITH GUANOSINE |
|---|
 | titl: the pnra (tp0319; tmpc) lipoprotein represents a new family of bacterial purine nucleoside receptor encoded within an atp-binding cassette (abc)-like operon in treponema pallidum |
| 3D3D | (1) | PNRA FROM TREPONEMA PALLIDUM COMPLEXED WITH ADENOSINE. |
|---|
 | titl: the pnra (tp0319; tmpc) lipoprotein represents a new family of bacterial purine nucleoside receptor encoded within an atp-binding cassette (abc)-like operon in treponema pallidum |
| 3D3D | (2) | ACTIVATED CONFORMATIONS OF THE RAS-GENE-ENCODED P21 PROTEIN. 1. AN ENERGY-REFINED STRUCTURE FOR THE NORMAL P21 PROTEIN COMPLEXED WITH GDP |
|---|
| 3D3D | (3) | HIGH-RESOLUTION STRUCTURE OF A PLASMID-ENCODED DIHYDROFOLATE REDUCTASE: PENTAGONAL NETWORK OF WATER MOLECULES IN THE D2-SYMMETRIC ACTIVE SITE |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF ECTROMELIA VIRUS EVM1 CHEMOKINE BINDING PROTEIN |
|---|
 | titl: structural determinants of chemokine binding by an ectromelia virus-encoded decoy receptor. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF CRYPTOSPORIDIUM PARVUM MALATE DEHYDROGENASE |
|---|
 | other_details: adjacent gene encodes predicted lactate dehydrogenase |
| 3D3D | (2) | SOLUTION STRUCTURE OF HUMAN P8-MTCP1, A CYSTEINE-RICH PROTEIN ENCODED BY THE MTCP1 ONCOGENE,REVEALS A NEW ALPHA-HELICAL ASSEMBLY MOTIF, NMR, 30 STRUCTURES |
|---|
| 3D3D | (1) | AMINOTRYPTOPHAN BARSTAR |
|---|
 | aminotryptophan, barstar, genetic code, protein folding, stability, hydrolase inhibitor |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE POLIOVIRUS PRECURSOR PROTEIN 3CD |
|---|
 | titl: crystal structure of poliovirus 3cd: virally-encoded protease and precursor to the rna-dependent rna polymerase. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE POLIOVIRUS RNA-DEPENDENT RNA POLYMERASE FIDELITY MUTANT 3DPOL G64S |
|---|
 | titl: crystal structure of poliovirus 3cd: virally-encoded protease and precursor to the rna-dependent rna polymerase. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PLAUTIA STALI INTESTINE VIRUS INTERGENIC REGION INTERNAL RIBOSOME ENTRY SITE RIBOSOMAL BINDING DOMAIN RNA AT 3.1 ANGSTROMS |
|---|
 | other_details: residues 6084, ins. code a-f, and 6119, ins. code a-e, are inserted to induce crystallization |
| 3D3D | (1) | STRUCTURE OF THE TYROSYL TRNA SYNTHETASE FROM ACANTHAMOEBA POLYPHAGA MIMIVIRUS COMPLEXED WITH TYROSYNOL |
|---|
 | titl: virus-encoded aminoacyl-trna synthetases: structural and functional characterization of mimivirus tyrrs and metrs. |
| 3D3D | (3) | CRYSTAL STRUCTURE OF THE UBIQUITIN-SPECIFIC PROTEASE ENCODED BY MURINE CYTOMEGALOVIRUS TEGUMENT PROTEIN M48 IN COMPLEX WITH A UBQUITIN-BASED SUICIDE SUBSTRATE |
|---|
| 3D3D | (1) | THE HYALURONAN BINDING DOMAIN OF MURINE CD44 |
|---|
 | other_details: encoded residues 25-174, equivalent to residues 1-151 of the mature protein, with additional residues m, n added at the n-terminus |
| 3D3D | (1) | THE HYALURONAN BINDING DOMAIN OF MURINE CD44 IN A TYPE A COMPLEX WITH AN HA 8-MER |
|---|
 | other_details: encoded residues 25-174, equivalent to residues 1-151 of the mature protein, with additional residues m, n added at the n-terminus |
| 3D3D | (1) | THE HYALURONAN BINDING DOMAIN OF MURINE CD44 IN A TYPE B COMPLEX WITH AN HA 8-MER |
|---|
 | other_details: encoded residues 25-174, equivalent to residues 1-151 of the mature protein, with additional residues m, n added at the n- terminus |
| 3D3D | (1) | SOLUTION STRUCTURE OF THE RGS DOMAIN FROM HUMAN RGS18 |
|---|
 | other_details: bl21 (de3) strain enriched with genes that encode rare trnas |
| 3D3D | (1) | SOLUTION STRUCTURE OF THE RGS DOMAIN OF HUMAN RGS14 |
|---|
 | other_details: bl21(de3) strain enriched with genes that encode rare trnas |
| 3D3D | (1) | SOLUTION STRUCTURE OF THE JOSEPHIN DOMAIN OF ATAXIN-3 IN COMPLEX WITH UBIQUITIN MOLECULE. |
|---|
 | titl: understanding the plasticity of the ubiquitin-protein recognition code: the josephin domain of ataxin-3 is a diubiquitin binding motif |
| 3D3D | (2) | NMR SOLUTION STRUCTURE OF THE PROTEIN CODED BY GENE RHOS4_12090 OF RHODOBACTER SPHAEROIDES. NORTHEAST STRUCTURAL GENOMICS TARGET RHR5 |
|---|
| 3D3D | (2) | SOLUTION NMR STRUCTURE OF PEFI (PLASMID-ENCODED FIMBRIAE REGULATORY) PROTEIN FROM SALMONELLA TYPHIMURIUM. NORTHEAST STRUCTURAL GENOMICS TARGET STR82 |
|---|
| 3D3D | (1) | NMR SOLUTION STRUCTURE OF A LDB1-LID:LHX3-LIM COMPLEX |
|---|
 | titl: implementing the lim code: the structural basis for cell type-specific assembly of lim-homeodomain complexes |
| 3D3D | (1) | NMR STRUCTURE OF THE GAMMA SUBUNIT OF CGMP PHOSPHODIESTERASE |
|---|
 | titl: intrinsically disordered gamma-subunit of cgmp phosphodiesterase encodes functionally relevant transient secondary and tertiary structure. |
| 3D3D | (1) | SOLUTION STRUCTURE OF COPK, A PERIPLASMIC PROTEIN INVOLVED IN COPPER RESISTANCE IN CUPRIAVIDUS METALLIDURANS CH34 |
|---|
 | copper, heavy metal resistance, open barrel, plasmid- encoded, metal binding protein |
| 3D3D | (2) | SOLUTION NMR STRUCTURE OF PROTEIN ENCODED BY GENE BPP1335 FROM BORDETELLA PARAPERTUSSIS: NORTHEAST STRUCTURAL GENOMICS TARGET BPR195 |
|---|
| 3D3D | (2) | SOLUTION NMR STRUCTURE OF PROTEIN ENCODED BY MTH693 FROM METHANOBACTERIUM THERMOAUTOTROPHICUM: NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT824A |
|---|
| 3D3D | (1) | GLYTM1B(1-19)ZIP: A CHIMERIC PEPTIDE MODEL OF THE N-TERMINUS OF A RAT SHORT ALPHA-TROPOMYOSIN WITH THE N-TERMINUS ENCODED BY EXON 1B IN COMPLEX WITH TM9D(252-284), A PEPTIDE MODEL CONTAINING THE C TERMINUS OF ALPHA-TROPOMYOSIN ENCODED BY EXON 9D |
|---|
| 3D3D | (1) | STRUCTURE OF TM1_TM2 IN LPPG MICELLES |
|---|
 | other_details: arabinose-induced e.coli vector, protein encoded as the delta-trp fusion, cleaved by cnbr cleavage |
| 3D3D | (1) | HUMAN JAGGED-1, EXON 6 |
|---|
 | titl: exon 6 of human jag1 encodes a conserved structural unit |
| 3D3D | (1) | NMR STRUCTURES OF GA95 AND GB95, TWO DESIGNED PROTEINS WITH 95% SEQUENCE IDENTITY BUT DIFFERENT FOLDS AND FUNCTIONS |
|---|
 | titl: a minimal sequence code for switching protein structure and function. |
| 3D3D | (1) | NMR STRUCTURES OF GA95 AND GB95, TWO DESIGNED PROTEINS WITH 95% SEQUENCE IDENTITY BUT DIFFERENT FOLDS AND FUNCTIONS |
|---|
 | titl: from the cover: a minimal sequence code for switching protein structure and function. |
| 3D3D | (1) | NMR SOLUTION STRUCTURE OF THE FIRST PHD FINGER DOMAIN OF HUMAN AUTOIMMUNE REGULATOR (AIRE) IN COMPLEX WITH HISTONE H3(1-20CYS) PEPTIDE |
|---|
| 3D3D | (2) | STRUCTURAL BASIS FOR HISTONE CODE RECOGNITION BY BRPF2-PHD1 FINGER |
|---|
 | phd finger, histone code, transcription | |
| structural basis for histone code recognition by brpf2-phd1 finger |
| 3D3D | (1) | COMPLEX BETWEEN BD1 OF BRD3 AND GATA-1 C-TAIL |
|---|
 | gata-1, brd3, histone code, nuclear protein-transcription complex |
| 3D3D | (1) | THIRD WW DOMAIN OF HUMAN NEDD4L IN COMPLEX WITH DOUBLY PHOSPHORYLATED HUMAN SMAD3 DERIVED PEPTIDE |
|---|
 | titl: a smad action turnover switch operated by ww domain readers of a phosphoserine code. |
| 3D3D | (1) | STRUCTURE OF THE SECOND WW DOMAIN FROM HUMAN YAP IN COMPLEX WITH A HUMAN SMAD1 DERIVED PEPTIDE |
|---|
 | titl: a smad action turnover switch operated by ww domain readers of a phosphoserine code. |
| 3D3D | (1) | STRUCTURE OF FIRST WW DOMAIN OF HUMAN YAP IN COMPLEX WITH A HUMAN SMAD1 DOUBLY-PHOSPHORILATED DERIVED PEPTIDE. |
|---|
 | titl: a smad action turnover switch operated by ww domain readers of a phosphoserine code. |
| 3D3D | (1) | STRUCTURE OF THE FIRST WW DOMAIN OF HUMAN YAP IN COMPLEX WITH A PHOSPHORYLATED HUMAN SMAD1 DERIVED PEPTIDE |
|---|
 | titl: a smad action turnover switch operated by ww domain readers of a phosphoserine code. |
| 3D3D | (1) | STRUCTURE OF THE FIRST WW DOMAIN OF HUMAN SMURF1 IN COMPLEX WITH A MONO-PHOSPHORYLATED HUMAN SMAD1 DERIVED PEPTIDE |
|---|
 | titl: a smad action turnover switch operated by ww domain readers of a phosphoserine code. |
| 3D3D | (1) | STRUCTURE OF THE FIRST WW DOMAIN OF HUMAN SMURF1 IN COMPLEX WITH A DI-PHOSPHORYLATED HUMAN SMAD1 DERIVED PEPTIDE |
|---|
 | titl: a smad action turnover switch operated by ww domain readers of a phosphoserine code. |
| 3D3D | (1) | STRUCTURE OF THE SECOND DOMAIN OF HUMAN SMURF1 IN COMPLEX WITH A HUMAN SMAD1 DERIVED PEPTIDE |
|---|
 | titl: a smad action turnover switch operated by ww domain readers of a phosphoserine code. |
| 3D3D | (1) | STRUCTURE OF THE SECOND DOMAIN OF HUMAN NEDD4L IN COMPLEX WITH A PHOSPHORYLATED PTPY MOTIF DERIVED FROM HUMAN SMAD3 |
|---|
 | titl: a smad action turnover switch operated by ww domain readers of a phosphoserine code. |
| 3D3D | (1) | STRUCTURE OF THE WW DOMAIN OF PIN1 IN COMPLEX WITH A HUMAN PHOSPHORYLATED SMAD3 DERIVED PEPTIDE |
|---|
 | titl: a smad action turnover switch operated by ww domain readers of a phosphoserine code. |
| 3D3D | (1) | SOLUTION STRUCTURE OF THE RMM-CTD DOMAINS OF HUMAN LINE-1 ORF1P |
|---|
 | molecule: orf1 codes for a 40 kda product |
| 3D3D | (2) | NOVEL DIMERIC STRUCTURE OF PHAGE PHI29-ENCODED PROTEIN P56: INSIGHTS INTO URACIL-DNA GLYCOSYLASE INHIBITION |
|---|
| 3D3D | (1) | ENTEROHAEMORRHAGIC E. COLI (EHEC) EXPLOITS A TRYPTOPHAN SWITCH TO HIJACK HOST F-ACTIN ASSEMBLY |
|---|
 | synonym: espf-like protein encoded on prophage u, tir-cytoskeleton coupling protein tccp |
| 3D3D | (2) | SOLUTION STRUCTURE OF HARMONIN N TERMINAL DOMAIN IN COMPLEX WITH A EXON68 ENCODED PEPTIDE OF CADHERIN23 |
|---|
 | fragment: exon68 encoded peptide | |
| solution structure of harmonin n terminal domain in complex with a exon68 encoded peptide of cadherin23 |
| 3D3D | (1) | SOLUTION STRUCTURE OF A CHAPERONE IN TYPE III SECRETION SYSTEM |
|---|
 | titl: substrate-activated conformational switch on chaperones encodes a targeting signal in type iii secretion. |
| 3D3D | (2) | THE BACTERIOPHAGE T7 ENCODED INHIBITOR (GP1.2) OF E. COLI DGTP TRIPHOSPHOHYDROLASE |
|---|
 | the bacteriophage t7 encoded inhibitor (gp1.2) of e. coli dgtp triphosphohydrolase | |
| titl: the bacteriophage t7 encoded inhibitor (gp1.2) of e. coli dgtp triphosphohydrolase |
| 3D3D | (1) | SOLUTION STRUCTURE OF THE C-TERMINALLY ENCODED PEPTIDE OF THE PLANT PARASITIC NEMATODE MELOIDOGYNE HAPLA - CEP11 |
|---|
 | solution structure of the c-terminally encoded peptide of the plant parasitic nematode meloidogyne hapla - cep11 |
| 3D3D | (1) | SOLUTION STRUCTURE OF THE C-TERMINALLY ENCODED PEPTIDE OF THE MODEL PLANT HOST MEDICAGO TRUNCATULA - CEP1 |
|---|
 | solution structure of the c-terminally encoded peptide of the model plant host medicago truncatula - cep1 |
| 3D3D | (1) | SOLUTION STRUCTURE OF THE CLAVATA ENCODED PEPTIDE OF ARABIDOPSIS THALIANA - ATCLE10 |
|---|
 | solution structure of the clavata encoded peptide of arabidopsis thaliana - atcle10 |
| 3D3D | (1) | SOLUTION STRUCTURE OF THE CLAVATA ENCODED PEPTIDE OF ARABIDOPSIS THALIANA - ATCLE44 |
|---|
 | solution structure of the clavata encoded peptide of arabidopsis thaliana - atcle44 |
| 3D3D | (2) | SOLUTION STRUCTURE OF THE CLAVATA-LIKE ENCODED PEPTIDE OF MELOIDOGYNE HAPLA - MHCLE4 |
|---|
 | solution structure of the clavata-like encoded peptide of meloidogyne hapla - mhcle4 | |
| molecule: clavata-like encoded peptide of meloidogyne hapla - mhcle4 |
| 3D3D | (2) | SOLUTION STRUCTURE OF THE CLAVATA-LIKE ENCODED PEPTIDE OF MELOIDOGYNE HAPLA - MHCLE5 |
|---|
 | molecule: clavata-like encoded peptide of meloidogyne hapla - mhcle5 | |
| solution structure of the clavata-like encoded peptide of meloidogyne hapla - mhcle5 |
| 3D3D | (2) | SOLUTION STRUCTURE OF THE CLAVATA-LIKE ENCODED PEPTIDE OF MELOIDOGYNE HAPLA - MHCLE6/7 |
|---|
 | solution structure of the clavata-like encoded peptide of meloidogyne hapla - mhcle6/7 | |
| molecule: clavata-like encoded peptide of meloidogyne hapla - mhcle6/7 |
| 3D3D | (1) | STF76 FROM THE SULFOLOBUS ISLANDICUS PLASMID-VIRUS PSSVX |
|---|
| 3D3D | (1) | SHIFTING THE POLARITY OF SOME CRITICAL RESIDUES IN MALARIAL PEPTIDES BINDING TO HOST CELLS IS A KEY FACTOR IN BREAKING CONSERVED ANTIGENS |
|---|
 | titl: shifting the polarity of some critical residues in malarial peptides' binding to host cells is a key factor in breaking conserved antigens' code of silence. |
| 3D3D | (1) | COMPLEX STRUCTURE OF DVL PDZ DOMAIN WITH LIGAND |
|---|
 | titl: protein-ligand interaction decoded by nmr chemical shift analysis |
| 3D3D | (2) | NMR STRUCTURE OF KORA, A PLASMID-ENCODED, GLOBAL TRANSCRIPTION REGULATOR KORA |
|---|
| 3D3D | (3) | ZIPCODE-BINDING-PROTEIN-1 KH3KH4(DD) DOMAINS IN COMPLEX WITH THE KH3 RNA TARGET |
|---|
| 3D3D | (3) | ZIPCODE-BINDING-PROTEIN-1 KH3(DD)KH4 DOMAINS IN COMPLEX WITH THE KH4 RNA TARGET |
|---|
| 3D3D | (1) | STRUCTURE OF THE 6TH ORF OF THE RHODOBACTER BLASTICA ATPASE OPERON; MAJASTRIDIN |
|---|
 | titl: crystal structure of a protein, structurally related to glycosyltransferases, encoded in the rhodobacter blasticus atp operon. |
| 3D3D | (1) | H98Q MUTANT OF THE HOMING ENDONUCLEASE I-PPOI COMPLEXED WITH DNA |
|---|
 | molecule: intron-encoded endonuclease i-ppoi |
| 3D3D | (1) | HUMAN EUCHROMATIC HISTONE METHYLTRANSFERASE 2 |
|---|
 | expression_system_strain: bl21(de3) code plus |
| 3D3D | (2) | STRUCTURE OF THE CONSERVED PROTEIN CODED BY LOCUS BT_0820 FROM BACTEROIDES THETAIOTAOMICRON |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF MONOBODY MBP-74/MALTOSE BINDING PROTEIN FUSION COMPLEX |
|---|
 | titl: high-affinity single-domain binding proteins with a binary-code interface. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF ASPARAGINE OXYGENASE IN COMPLEX WITH FE(II) |
|---|
 | other_details: expression plasmid encodes for n-terminal his-7-tag for imac |
| 3D3D | (1) | CYSTAL STRUCTURE OF ASPARAGINE OXYGENASE IN COMPLEX WITH FE(II), 2S, 3S-3-HYDROXYASPARAGINE AND SUCCINATE |
|---|
 | other_details: expression plasmid encodes for n-terminal his-7-tag for imac |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE MIMIVIRUS CYCLOPHILIN |
|---|
 | titl: structural, biochemical, and in vivo characterization of the first virally encoded cyclophilin from the mimivirus. |
| 3D3D | (1) | 13-DEOXYTEDANOLIDE BOUND TO THE LARGE SUBUNIT OF HALOARCULA MARISMORTUI |
|---|
 | other_details: encoded by rrna operons |
| 3D3D | (1) | GIRODAZOLE BOUND TO THE LARGE SUBUNIT OF HALOARCULA MARISMORTUI |
|---|
 | other_details: encoded by rrna operons |
| 3D3D | (1) | SOLUTION STRUCTURE OF THE RGS DOMAIN FROM HUMAN RGS18 |
|---|
 | other_details: bl21 (de3) strain enriched with genes that encode rare trnas |
| 3D3D | (1) | A CRYSTAL STRUCTURE OF PPAR ALPHA BOUND WITH SRC1 PEPTIDE AND GW735 |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF A GLYCOSYLTRANSFERASE INVOLVED IN THE GLYCOSYLATION OF THE MAJOR CAPSID OF PBCV-1 |
|---|
 | titl: structure and function of a chlorella virus-encoded glycosyltransferase. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A GLYCOSYLTRANSFERASE INVOLVED IN THE GLYCOSYLATION OF THE MAJOR CAPSID OF PBCV-1 |
|---|
 | titl: structure and function of a chlorella virus-encoded glycosyltransferase. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A GLYCOSYLTRANSFERASE INVOLVED IN THE GLYCOSYLATION OF THE MAJOR CAPSID OF PBCV-1 |
|---|
 | titl: structure and function of a chlorella virus-encoded glycosyltransferase. |
| 3D3D | (2) | CRYSTAL STRUCTURE OF GENOMICALLY ENCODED FOSFOMYCIN RESISTANCE PROTEIN, FOSX, FROM LISTERIA MONOCYTOGENES (HEXAGONAL FORM) |
|---|
| 3D3D | (2) | CRYSTAL STRUCTURE OF MONOCLINIC FORM OF GENOMICALLY ENCODED FOSFOMYCIN RESISTANCE PROTEIN, FOSX, FROM LISTERIA MONOCYTOGENES AT PH 5.75 |
|---|
| 3D3D | (2) | CRYSTAL STRUCTURE OF MONOCLINIC FORM OF GENOMICALLY ENCODED FOSFOMYCIN RESISTANCE PROTEIN, FOSX, FROM LISTERIA MONOCYTOGENES AT PH 6.5 |
|---|
| 3D3D | (2) | CRYSTAL STRUCTURE OF GENOMICALLY ENCODED FOSFOMYCIN RESISTANCE PROTEIN, FOSX, FROM LISTERIA MONOCYTOGENES (TETRAGONAL FORM) |
|---|
| 3D3D | (2) | CRYSTAL STRUCTURE OF GENOMICALLY ENCODED FOSFOMYCIN RESISTANCE PROTEIN, FOSX, FROM LISTERIA MONOCYTOGENES COMPLEXED WITH MN(II) AND SULFATE ION |
|---|
| 3D3D | (2) | CRYSTAL STRUCTURE OF E126Q MUTANT OF GENOMICALLY ENCODED FOSFOMYCIN RESISTANCE PROTEIN, FOSX, FROM LISTERIA MONOCYTOGENES COMPLEXED WITH MN(II) AND 1S,2S-DIHYDROXYPROPYLPHOSPHONIC ACID |
|---|
| 3D3D | (1) | ARG-MODIFIED HUMAN BETA-DEFENSIN 1 (HBD1) |
|---|
 | titl: toward understanding the cationicity of defensins. arg and lys versus their noncoded analogs. |
| 3D3D | (1) | HUMAN ALPHA-DEFENSIN 1 (MULTIPLE ARG->LYS MUTANT) |
|---|
 | titl: toward understanding the cationicity of defensins. arg and lys versus their noncoded analogs. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE BHC80 PHD FINGER |
|---|
 | phd finger, histone code, braf-hdac complex, transcription |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A BIPYRIDYLALANYL-TRNA SYNTHETASE |
|---|
 | titl: a genetically encoded bidentate, metal-binding amino acid. |
| 3D3D | (1) | HUMAN PUTATIVE METHYLTRANSFERASE MGC2408 |
|---|
 | titl: an intact sam-dependent methyltransferase fold is encoded by the human endothelin-converting enzyme-2 gene. |
| 3D3D | (1) | CRYSTAL STRUCTURES OF IMMUNE RECEPTOR COMPLEXES |
|---|
 | titl: structural evidence for a germline-encoded t cell receptor-major histocompatibility complex interaction 'codon'. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF IMAZG FROM VIBRIO DAT 722: NTAG-IMAZG (P43212) |
|---|
 | titl: a putative house-cleaning enzyme encoded within an integron array: 1.8 a crystal structure defines a new mazg subtype. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF IMAZG FROM VIBRIO DAT 722: CTAG-IMAZG (P41212) |
|---|
 | titl: a putative house-cleaning enzyme encoded within an integron array: 1.8 a crystal structure defines a new mazg subtype. |
| 3D3D | (1) | THE STRUCTURE OF PYRROLYSYL-TRNA SYNTHETASE BOUND TO AN ATP ANALOGUE |
|---|
 | titl: structure of pyrrolysyl-trna synthetase, an archaeal enzyme for genetic code innovation. |
| 3D3D | (1) | PYRROLYSINE TRNA SYNTHETASE BOUND TO A PYRROLYSINE ANALOGUE (CYC) AND ATP |
|---|
 | titl: structure of pyrrolysyl-trna synthetase, an archaeal enzyme for genetic code innovation. |
| 3D3D | (1) | PYRROLYSYL-TRNA SYNTHETASE BOUND TO ADENYLATED PYRROLYSINE AND PYROPHOSPHATE |
|---|
 | titl: structure of pyrrolysyl-trna synthetase, an archaeal enzyme for genetic code innovation. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF IMAZG FROM VIBRIO DAT 722: CTAG-IMAZG (P43212) |
|---|
 | titl: a putative house-cleaning enzyme encoded within an integron array: 1.8 a crystal structure defines a new mazg subtype. |
| 3D3D | (1) | THE UDP COMPLEX STRUCTURE OF THE SIXTH GENE PRODUCT OF THE F1-ATPASE OPERON OF RHODOBACTER BLASTICUS |
|---|
 | titl: crystal structure of a protein, structurally related to glycosyltransferases, encoded in the rhodobacter blasticus atp operon. |
| 3D3D | (1) | BIFUNCTIONAL DCTP DEAMINASE:DUTPASE FROM MYCOBACTERIUM TUBERCULOSIS, APO FORM |
|---|
 | titl: mechanism of dttp inhibition of the bifunctional dctp deaminase:dutpase encoded by mycobacterium tuberculosis |
| 3D3D | (1) | COEVOLUTION OF A HOMING ENDONUCLEASE AND ITS HOST TARGET SEQUENCE |
|---|
 | molecule: intron-encoded dna endonuclease i-anii |
| 3D3D | (1) | THE CRYSTAL STRUCTURE OF THE E.COLI ESPP AUTOTRANSPORTER BETA-DOMAIN. |
|---|
 | synonym: extracellular serine protease plasmid-encoded espp |
| 3D3D | (1) | BIFUNCTIONAL DCTP DEAMINASE: DUTPASE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH DTTP |
|---|
 | titl: mechanism of dttp inhibition of the bifunctional dctp deaminase:dutpase encoded by mycobacterium tuberculosis. |
| 3D3D | (2) | FUNCTIONAL ANNOTATION OF ESCHERICHIA COLI YIHS-ENCODED PROTEIN |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF LHX3 LIM DOMAINS 1 AND 2 WITH THE BINDING DOMAIN OF ISL1 |
|---|
 | titl: implementing the lim code: the structural basis for cell type-specific assembly of lim-homeodomain complexes. |
| 3D3D | (1) | HIGH RESOLUTION DHFR R-67 |
|---|
 | folate metabolism, plasmid-encoded r67 dhfr, tmp-resistant dhfr, antibiotic resistance, methotrexate resistance, nadp, one-carbon metabolism, oxidoreductase, trimethoprim resistance |
| 3D3D | (1) | SYNTHETIC GENE ENCODED BACILLUS SUBTILIS FTSZ WITH BOUND SULFATE ION |
|---|
 | synthetic gene encoded bacillus subtilis ftsz with bound sulfate ion |
| 3D3D | (1) | SYNTHETIC GENE ENCODED BACILLUS SUBTILIS FTSZ WITH TWO SULFATE IONS AND SODIUM ION IN THE NUCLEOTIDE POCKET |
|---|
 | synthetic gene encoded bacillus subtilis ftsz with two sulfate ions and sodium ion in the nucleotide pocket |
| 3D3D | (1) | SYNTHETIC GENE ENCODED BACILLUS SUBTILIS FTSZ NCS DIMER WITH BOUND GDP |
|---|
 | synthetic gene encoded bacillus subtilis ftsz ncs dimer with bound gdp |
| 3D3D | (1) | SYNTHETIC GENE ENCODED BACILLUS SUBTILIS FTSZ NCS DIMER WITH BOUND GDP AND GTP-GAMMA-S |
|---|
 | synthetic gene encoded bacillus subtilis ftsz ncs dimer with bound gdp and gtp-gamma-s |
| 3D3D | (1) | STRUCTURE OF CRME, A POXVIRUS TNF RECEPTOR |
|---|
 | titl: structure of crme, a virus-encoded tumour necrosis factor receptor. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF COACTIVATOR-ASSOCIATED ARGININE METHYLTRANSFERASE 1 (CARM1), IN COMPLEX WITH S-ADENOSYL-HOMOCYSTEINE |
|---|
 | titl: insights into histone code syntax from structural and biochemical studies of carm1 methyltransferase |
| 3D3D | (1) | CRYSTAL STRUCTURE OF COACTIVATOR-ASSOCIATED ARGININE METHYLTRANSFERASE 1 (CARM1), UNLIGANDED |
|---|
 | titl: insights into histone code syntax from structural and biochemical studies of carm1 methyltransferase |
| 3D3D | (1) | CRYSTAL STRUCTURE OF ALDEHYDE DEHYDROGENASE FROM BURKHOLDERIA XENOVORANS LB400 |
|---|
 | titl: structural and biochemical characterization of a novel aldehyde dehydrogenase encoded by the benzoate oxidation (box) pathway in burkholderia xenovorans lb400 |
| 3D3D | (1) | STRUCTURE OF A STREPTOCOCCUS MUTANS CE4 ESTERASE |
|---|
 | titl: streptococcus mutans smu.623c codes for a functional, metal dependent polysaccharide deacetylase that modulates interactions with salivary agglutinin. |
| 3D3D | (2) | STRUCTURE OF THE HUMAN LINE-1 ORF1P CENTRAL DOMAIN |
|---|
 | synonym: orf1 codes for a 40 kda product | |
| titl: non-ltr retrotransposons encode noncanonical rrm domains in their first open reading frame. |
| 3D3D | (1) | THE PARTIAL STRUCTURE OF A GROUP A STREPTOCOCCAL PHAGE-ENCODED TAIL FIBRE HYALURONATE LYASE HYLP3 |
|---|
 | the partial structure of a group a streptococcal phage-encoded tail fibre hyaluronate lyase hylp3 |
| 3D3D | (1) | PT26-6P |
|---|
 | titl: a protein encoded by a new family of mobile elements from euryarchaea exhibits three domains with novel folds. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE OXA-10 V117T MUTANT AT PH 6.5 INHIBITED BY A CHLORIDE ION |
|---|
 | antibiotic resistance, plasmid encoded, hydrolase |
| 3D3D | (1) | THE PARTIAL STRUCTURE OF A GROUP A STREPTPCOCCAL PHAGE-ENCODED TAIL FIBRE HYALURONATE LYASE HYLP2 |
|---|
 | the partial structure of a group a streptpcoccal phage-encoded tail fibre hyaluronate lyase hylp2 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE ACYL-ENZYME OXA-10 K70C-AMPICILLIN AT PH 7 |
|---|
 | hydrolase, antibiotic resistance, class d beta-lactamase, plasmid encoded |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE OXA-10 K70C MUTANT AT PH 7.0 |
|---|
 | class d, plasmid encoded, hydrolase, antibiotic resistance, beta-lactamase |
| 3D3D | (1) | STRUCTURE OF A PHOTOACTIVATABLE RAC1 CONTAINING LOV2 WILDTYPE |
|---|
 | titl: a genetically encoded photoactivatable rac controls the motility of living cells. |
| 3D3D | (1) | STRUCTURE OF A PHOTOACTIVATABLE RAC1 CONTAINING THE LOV2 C450A MUTANT |
|---|
 | titl: a genetically encoded photoactivatable rac controls the motility of living cells. |
| 3D3D | (1) | STRUCTURE OF A PHOTOACTIVATABLE RAC1 CONTAINING THE LOV2 C450M MUTANT |
|---|
 | titl: a genetically encoded photoactivatable rac controls the motility of living cells. |
| 3D3D | (1) | EXTRACELLULAR ENDONUCLEASE |
|---|
 | titl: the structural characterization of a prophage- encoded extracellular dnase from streptococcus pyogenes. |
| 3D3D | (1) | EXTRACELLULAR NUCLEASE |
|---|
 | titl: the structural characterization of a prophage- encoded extracellular dnase from streptococcus pyogenes. |
| 3D3D | (2) | CRYSTAL STRUCTURE OF E167A MUTANT OF THE BOX PATHWAY ENCODED ALDH FROM BURKHOLDERIA XENOVORANS LB400 |
|---|
| 3D3D | (2) | CRYSTAL STRUCTURE OF E496A MUTANT OF THE BOX PATHWAY ENCODED ALDH FROM BURKHOLDERIA XENOVORANS LB400 |
|---|
| 3D3D | (2) | CRYSTAL STRUCTURE OF E257Q MUTANT OF THE BOX PATHWAY ENCODED ALDH FROM BURKHOLDERIA XENOVORANS LB400 |
|---|
| 3D3D | (2) | CRYSTAL STRUCTURE OF C296A MUTANT OF THE BOX PATHWAY ENCODED ALDH FROM BURKHOLDERIA XENOVORANS LB400 |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD2 WITH THE INHIBITOR GW841819X |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD2 WITH THE INHIBITOR GSK525762 (IBET) |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 WITH THE INHIBITOR GW841819X |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE SECOND BROMODOMAIN OF HUMAN BRD4 WITH THE INHIBITOR GW841819X |
|---|
| 3D3D | (2) | A SPECIFIC AND MODULAR BINDING CODE FOR CYTOSINE RECOGNITION IN PUF DOMAINS |
|---|
| 3D3D | (1) | STRUCTURE OF THE HUMAN LINE-1 ORF1P TRIMER |
|---|
 | synonym: orf1 codes for a 40 kda product |
| 3D3D | (1) | STRUCTURE OF THE HUMAN LINE-1 ORF1P TRIMER |
|---|
 | synonym: orf1 codes for a 40 kda product |
| 3D3D | (1) | STRUCTURE OF THE HUMAN LINE-1 ORF1P TRIMER |
|---|
 | synonym: orf1 codes for a 40 kda product |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HYLURANIDASE TRIMER AT 2.6 A RESOLUTION |
|---|
 | titl: polysaccharide binding sites in hyaluronate lyase--crystal structures of native phage-encoded hyaluronate lyase and it complexes with ascorbic acid and lactose |
| 3D3D | (1) | STRUCTUAL BASIS OF AZIDO-TYROSINE RECOGNITION BY ENGINEERED BACTERIAL TYROSYL-TRNA SYNTHETASE |
|---|
 | titl: functional replacement of the endogenous tyrosyl-trna synthetase-trnatyr pair by the archaeal tyrosine pair in escherichia coli for genetic code expansion |
| 3D3D | (1) | CRYSTAL STRUCTURE OF IMMUNE RECEPTOR COMPLEX |
|---|
 | titl: structural evidence for a germline-encoded t cell receptor-major histocompatibility complex interaction 'codon' |
| 3D3D | (1) | CRYSTAL STRUCTURE OF IMMUNE RECEPTOR |
|---|
 | titl: structural evidence for a germline-encoded t cell receptor-major histocompatibility complex interaction 'codon' |
| 3D3D | (1) | CRYSTAL STRUCTURE OF FFRP-DM1 |
|---|
 | titl: a structural code for discriminating between transcription signals revealed by the feast/famine regulatory protein dm1 in complex with ligands |
| 3D3D | (2) | FUNCTIONAL ANNOTATION OF SALMONELLA ENTERICA YIHS-ENCODED PROTEIN |
|---|
| 3D3D | (1) | PYRROLYSYL-TRNA SYNTHETASE BOUND TO ADENYLATED PYRROLYSINE AND PYROPHOSPHATE |
|---|
 | titl: structure of pyrrolysyl-trna synthetase, an archaeal enzyme for genetic code innovation. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF PYRROLYSYL-TRNA SYNTHETASE IN COMPLEX WITH BOCLYS AND AN ATP ANALOGUE |
|---|
 | titl: multistep engineering of pyrrolysyl-trna synthetase to genetically encode n(varepsilon)-(o-azidobenzyloxycarbonyl) lysine for site-specific protein modification |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF PYRROLYSYL-TRNA SYNTHETASE IN COMPLEX WITH ALOCLYS-AMP AND PNP |
|---|
 | titl: multistep engineering of pyrrolysyl-trna synthetase to genetically encode n(varepsilon)-(o-azidobenzyloxycarbonyl) lysine for site-specific protein modification |
| 3D3D | (1) | STRUCTURE OF GANKYRIN-S6ATPASE PHOTO-CROSS-LINKED SITE-SPECIFICALLY, AND INCOPORATED BY GENETIC CODE EXPANSION |
|---|
 | structure of gankyrin-s6atpase photo-cross-linked site-specifically, and incoporated by genetic code expansion |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE HUMAN SNP PAD4 PROTEIN |
|---|
 | titl: structural and biochemical analyses of the human pad4 variant encoded by a functional haplotype gene |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE HUMAN WILD-TYPE PAD4 PROTEIN |
|---|
 | titl: structural and biochemical analyses of the human pad4 variant encoded by a functional haplotype gene |
| 3D3D | (1) | SYNTHETIC GENE ENCODED DCPS BOUND TO INHIBITOR DG156844 |
|---|
 | synthetic gene encoded dcps bound to inhibitor dg156844 |
| 3D3D | (1) | SYNTHETIC GENE ENCODED DCPS BOUND TO INHIBITOR DG157493 |
|---|
 | synthetic gene encoded dcps bound to inhibitor dg157493 |
| 3D3D | (1) | SYNTHETIC GENE ENCODED DCPS BOUND TO INHIBITOR DG153249 |
|---|
 | synthetic gene encoded dcps bound to inhibitor dg153249 |
| 3D3D | (1) | STRUCTURE OF A CHONDROITIN SULPHATE BINDING DBL3X FROM A VAR2CSA ENCODED PFEMP1 PROTEIN |
|---|
 | structure of a chondroitin sulphate binding dbl3x from a var2csa encoded pfemp1 protein |
| 3D3D | (2) | STRUCTURE OF A CHONDROITIN SULPHATE BINDING DBL3X FROM A VAR2CSA ENCODED PFEMP1 PROTEIN IN COMPLEX WITH SULPHATE |
|---|
| 3D3D | (2) | STRUCTURE OF A CHONDROITIN SULPHATE BINDING DBL3X DOMAIN FROM A VAR2CSA ENCODED PFEMP1 PROTEIN |
|---|
| 3D3D | (1) | I-SCEI IN COMPLEX WITH A BOTTOM NICKED DNA SUBSTRATE |
|---|
 | molecule: intron-encoded endonuclease i-scei |
| 3D3D | (1) | I-SCEI IN COMPLEX WITH A TOP NICKED DNA SUBSTRATE |
|---|
 | molecule: intron-encoded endonuclease i-scei |
| 3D3D | (2) | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED PROTEIN ENCODED BY CRYPTIC PROPHAGE |
|---|
 | crystal structure of an uncharacterized protein encoded by cryptic prophage | |
| titl: crystal structure of an uncharacterized protein encoded by cryptic prophage. |
| 3D3D | (1) | THE TPR DOMAIN OF HUMAN KINESIN LIGHT CHAIN 2 (HKLC2) |
|---|
 | expression_system_strain: bl21coden(+) |
| 3D3D | (1) | CRYSTAL STRUCTURE OF MALATE DEHYDROGENASE FROM BURKHOLDERIA PSEUDOMALLEI |
|---|
 | ssgcid, structural genomics, niaid, decode biostructures, malate dehydrogenase, nad, oxidoreductase, tricarboxylic acid cycle, seattle structural genomics center for infectious disease |
| 3D3D | (1) | CRYSTAL STRUCTURE OF 4-(TRIFLUOROMETHYLDIAZIRINYL) PHENYLALANYL-TRNA SYNTHETASE |
|---|
 | titl: a genetically encoded diazirine photocrosslinker in escherichia coli |
| 3D3D | (1) | CRYSTAL STRUCTURE OF 4-(TRIFLUOROMETHYLDIAZIRINYL) PHENYLALANYL-TRNA SYNTHETASE |
|---|
 | titl: a genetically encoded diazirine photocrosslinker in escherichia coli |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE REPLICATION INITIATOR PROTEIN ENCODED ON PLASMID PMV158 (REPB), TRIGONAL FORM, TO 2.7 ANG RESOLUTION |
|---|
 | crystal structure of the replication initiator protein encoded on plasmid pmv158 (repb), trigonal form, to 2.7 ang resolution |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE REPLICATION INITIATOR PROTEIN ENCODED ON PLASMID PMV158 (REPB), TETRAGONAL FORM, TO 3.6 ANG RESOLUTION |
|---|
 | crystal structure of the replication initiator protein encoded on plasmid pmv158 (repb), tetragonal form, to 3.6 ang resolution |
| 3D3D | (1) | CRYSTAL STRUCTURE OF NAD+ SYNTHETASE FROM BURKHOLDERIA PSEUDOMALLEI |
|---|
 | ssgcid, decode, structural genomics, psi, protein structure initiative, seattle structural genomics center for infectious disease, ligase |
| 3D3D | (2) | ARCHAEAL INTRON-ENCODED HOMING ENDONUCLEASE I-VDI141I COMPLEXED WITH DNA |
|---|
 | archaeal intron-encoded homing endonuclease i-vdi141i complexed with dna | |
| molecule: rrna intron-encoded endonuclease |
| 3D3D | (1) | CRYSTAL STRUCTURE OF TRMH FAMILY RNA METHYLTRANSFERASE FROM BURKHOLDERIA PSEUDOMALLEI |
|---|
 | ssgcid, protein knot, decode, rna methyltransferase, methyltransferase, transferase, structural genomics, seattle structural genomics center for infectious disease |
| 3D3D | (1) | CRYSTAL STRUCTURE OF 3-OXOACYL-(ACYL CARRIER PROTEIN) SYNTHASE II FROM BARTONELLA HENSELAE |
|---|
 | ssgcid, decode, transferase, structural genomics, seattle structural genomics center for infectious disease |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PRECORRIN-8X METHYL MUTASE CBIC/COBH FROM BRUCELLA MELITENSIS |
|---|
 | ssgcid, decode, braba00006a, structural genomics, seattle structural genomics center for infectious disease, isomerase |
| 3D3D | (1) | CRYSTAL STRUCTURE OF SERINE HYDROXYMETHYLTRANSFERASE FROM BURKHOLDERIA PSEUDOMALLEI |
|---|
 | ssgcid, decode, bupsa00008a, one-carbon metabolism, pyridoxal phosphate, transferase, structural genomics, seattle structural genomics center for infectious disease |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE MUTATED S328N HKLC2 TPR DOMAIN |
|---|
 | expression_system_strain: bl21 coden(+) |
| 3D3D | (1) | CRYSTAL STRUCTURE OF Y2 I-ANII VARIANT (F13Y/S111Y)/DNA COMPLEX WITH CALCIUM |
|---|
 | molecule: intron-encoded dna endonuclease i-anii |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE COMPLEX OF HYALURANIDASE TRIMER WITH ASCORBIC ACID AT 3.1 A RESOLUTION REVEALS THE LOCATIONS OF THREE BINDING SITES |
|---|
 | titl: polysaccharide binding sites in hyaluronate lyase--crystal structures of native phage-encoded hyaluronate lyase and its complexes with ascorbic acid and lactose. |
| 3D3D | (1) | AILSST SEGMENT FROM ISLET AMYLOID POLYPEPTIDE |
|---|
 | titl: molecular mechanisms for protein-encoded inheritance. |
| 3D3D | (1) | HSSNNF SEGMENT FROM ISLET AMYLOID POLYPEPTIDE (IAPP OR AMYLIN) |
|---|
 | titl: molecular mechanisms for protein-encoded inheritance. |
| 3D3D | (1) | NFLVHS SEGMENT FROM ISLET AMYLOID POLYPEPTIDE (IAPP OR AMYLIN) |
|---|
 | titl: molecular mechanisms for protein-encoded inheritance |
| 3D3D | (1) | NFLVHSS SEGMENT FROM ISLET AMYLOID POLYPEPTIDE (IAPP OR AMYLIN) |
|---|
 | titl: molecular mechanisms for protein-encoded inheritance |
| 3D3D | (1) | NVGSNTY SEGMENT FROM ISLET AMYLOID POLYPEPTIDE (IAPP OR AMYLIN), HYDRATED CRYSTAL FORM |
|---|
 | titl: molecular mechanisms for protein-encoded inheritance |
| 3D3D | (1) | NVGSNTY SEGMENT FROM ISLET AMYLOID POLYPEPTIDE (IAPP OR AMYLIN), DEHYDRATED CRYSTAL FORM |
|---|
 | titl: molecular mechanisms for protein-encoded inheritance |
| 3D3D | (1) | STRUCTURE OF AN AMYLOID FORMING PEPTIDE SSTNVG FROM IAPP (ALTERNATE POLYMORPH) |
|---|
 | titl: molecular mechanisms for protein-encoded inheritance. |
| 3D3D | (1) | NNQNTF SEGMENT FROM ELK PRION |
|---|
 | titl: molecular mechanisms for protein-encoded inheritance. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF FERRITIN (BACTERIOFERRITIN) FROM BRUCELLA MELITENSIS |
|---|
 | niaid, ssgcid, decode, ferritin, structural genomics, seattle structural genomics center for infectious disease, metal binding protein |
| 3D3D | (2) | CRYSTAL STRUCTURE OF PUTATIVE DNA MODIFICATION METHYLTRANSFERASE ENCODED WITHIN PROPHAGE CP-933R (E.COLI) |
|---|
| 3D3D | (1) | CCDB DIMER IN COMPLEX WITH TWO C-TERMINAL CCDA DOMAINS |
|---|
 | other_details: fragment of the ccda protein encoded on f- plasmid. synthesised via solid phase syntesis. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF MALONYL COA-ACYL CARRIER PROTEIN TRANSACYLASE FROM BURKHOLDERIA PSEUDOMALLEI USING DRIED SEAWEED AS NUCLEANT OR PROTEASE |
|---|
 | ssgcid, niaid, decode biostructures, dried seaweed, acyltransferase, transferase, structural genomics, seattle structural genomics center for infectious disease |
| 3D3D | (3) | X-RAY STRUCTURE OF GENETICALLY ENCODED PHOTOSENSITIZER KILLERRED IN NATIVE FORM |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF PYRIDOXAL PHOSPHATE BIOSYNTHETIC PROTEIN FROM BURKHOLDERIA PSEUDOMALLEI |
|---|
 | decode, ssgcid, niaid, sbri, pyridoxine biosynthesis, transferase, structural genomics, seattle structural genomics center for infectious disease |
| 3D3D | (1) | CRYSTAL STRUCTURE OF N-ETHYLMALEIMIDINE REDUCTASE FROM BURKHOLDERIA PSEUDOMALLEI |
|---|
 | decode biostructures, ssgcid, niaid, targetdb bupsa00093a, structural genomics, seattle structural genomics center for infectious disease, oxidoreductase |
| 3D3D | (2) | X-RAY STRUCTURE OF PHOTOBLEACHED KILLERRED |
|---|
 | titl: structural basis for phototoxicity of the genetically encoded photosensitizer killerred. | |
| fluorescent protein, genetically encoded photosensitizer, killerred, phototoxicity |
| 3D3D | (1) | CRYSTAL STRUCTURE OF NICOTINATE-NUCLEOTIDE PYROPHOSPHORYLASE FROM BURKHOLDERI PSEUDOMALLEI |
|---|
 | decode biostructures, ssgcid, niaid, sbri, uwppg, glycosyltransferase, transferase, structural genomics, seattle structural genomics center for infectious disease |
| 3D3D | (1) | CRYSTAL STRUCTURE OF GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, TYPE I FROM BURKHOLDERIA PSEUDOMALLEI |
|---|
 | decode biostructures, ssgcid, uwppg, sbri, niaid, oxidoreductase, structural genomics, seattle structural genomics center for infectious disease |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PHOSPHOGLYCEROMUTASE FROM BURKHOLDERIA PSEUDOMALLEI WITH 2-PHOSPHOSERINE |
|---|
 | phosphoglyceromutase, decode, sbri, niaid, uwppg, glycolysis, isomerase, structural genomics, seattle structural genomics center for infectious disease, ssgcid |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PHOSPHOGLYCEROMUTASE FROM BURKHOLDERIA PSEUDOMALLEI WITH 3-PHOSPHOGLYCERIC ACID AND VANADATE |
|---|
 | phosphoglyceromutase, decode, uwppg. sbri, niaid, ssgcid, glycolysis, isomerase, structural genomics, seattle structural genomics center for infectious disease |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PROBABLE SHORT-CHAIN DEHYDROGENASE-REDUCTASE FROM MYCOBACTERIUM TUBERCULOSIS |
|---|
 | ssgcid, decode, niaid, uwppg, sbri, oxidoreductase, structural genomics, seattle structural genomics center for infectious disease |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PHOSPHOGLYCEROMUTASE FROM BURKHOLDERIA PSEUDOMALLEI WITH VANADATE AND GLYCEROL |
|---|
 | ssgcid, niaid, decode, uwppg, sbri, glycolysis, isomerase, structural genomics, seattle structural genomics center for infectious disease |
| 3D3D | (1) | STRUCTURE OF ARTA AND DNA COMPLEX |
|---|
 | titl: the staphylococcus aureus psk41 plasmid-encoded arta protein is a master regulator of plasmid transmission genes and contains a rhh motif used in alternate dna-binding modes. |
| 3D3D | (1) | ALGAL PROLYL 4-HYDROXYLASE COMPLEXED WITH ZINC AND (SER-PRO)5 PEPTIDE SUBSTRATE |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF PRIMASE REPB' |
|---|
 | titl: structure and function of primase repb' encoded by broad-host-range plasmid rsf1010 that replicates exclusivel in leading-strand mode |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF PRIMASE REPB' IN COMPLEX WITH INITIATOR DNA |
|---|
 | titl: structure and function of primase repb' encoded by broad-host-range plasmid rsf1010 that replicates exclusivel in leading-strand mode |
| 3D3D | (1) | CRYSTAL STRUCTURE OF ENOYL-COA HYDRATASE FROM MYCOBACTERIUM TUBERCULOSIS |
|---|
 | niaid, decode, infectious disease, mpcs, fatty acid metabolism, lipid metabolism, lyase, structural genomics, seattle structural genomics center for infectious disease, ssgcid |
| 3D3D | (1) | CRYSTAL STRUCTURE OF AURORA A IN COMPLEX WITH TPX2 AND COMPOUND 10 |
|---|
 | titl: design, synthesis and selection of dna-encoded small-molecule libraries. |
| 3D3D | (1) | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/COMPOUND 14B |
|---|
 | titl: design, synthesis and selection of dna-encoded small-molecule libraries. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF GLYCINE CLEAVAGE SYSTEM PROTEIN H FROM MYCOBACTERIUM TUBERCULOSIS |
|---|
 | ssgcid, niaid, decode, uw, sbri, lipoyl, structural genomics, seattle structural genomics center for infectious disease, unknown function, oxidoreductase |
| 3D3D | (1) | CRYSTAL STRUCTURE OF RIBOSE-5-PHOSPHATE ISOMERASE A FROM BARTONELLA HENSELAE |
|---|
 | niaid, ssgcid, decode, sbri, uw, bartonella, isomerase, structural genomics, seattle structural genomics center for infectious disease |
| 3D3D | (1) | CRYSTAL STRUCTURE OF MUTATOR MUTT FROM BARTONELLA HENSELAE |
|---|
 | niaid, ssgcid, decode, uw, sbri, infectious diseases, hydrolase, structural genomics, seattle structural genomics center for infectious disease |
| 3D3D | (1) | CRYSTAL STRUCTURE OF GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM BORRELIA BURGDORFERI |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF PROBABLE THIOESTERASE FROM BARTONELLA HENSELAE |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF GLUCOSAMINE-6-PHOSPHATE DEAMINASE FROM BORRELIA BURGDORFERI |
|---|
| 3D3D | (1) | CCDB DIMER IN COMPLEX WITH ONE C-TERMINAL CCDA DOMAIN |
|---|
 | other_details: fragment of the ccda protein encoded on f- plasmid. synthesised via solid phase synthesis by bio- synthesis. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE CATALYTIC FRAGMENT OF ALANYL-TRNA SYNTHETASE IN COMPLEX WITH L-SERINE: RE-REFINED |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF PROBABLE THIOSULFATE SULFURTRANSFERASE CYSA2 (RHODANESE-LIKE PROTEIN) FROM MYCOBACTERIUM TUBERCULOSIS |
|---|
 | niaid, ssgcid, seattle structural center for infectious disease, decode, uw, sbri, tuberculosis, transferase, structural genomics, seattle structural genomics center for infectious disease |
| 3D3D | (1) | CRYSTAL STRUCTURE OF METHYLCITRATE SYNTHASE FROM MYCOBACTERIUM TUBERCULOSIS |
|---|
 | niaid, ssgcid, structural genomics, seattle structural genomics center for infectious disease, tubercluosis, uw, sbri, decode, acyltransferase, transferase |
| 3D3D | (1) | CRYSTAL STRUCTURE OF T4 LYSOZYME WITH THE UNNATURAL AMINO ACID P-ACETYL-L-PHENYLALANINE INCORPORATED AT POSITION 131 |
|---|
 | titl: site-directed spin labeling of a genetically encoded unnatural amino acid. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF XYLULOKINASE FROM CHROMOBACTERIUM VIOLACEUM |
|---|
 | expression_system_strain: bl21(de3)-coden+ril-stratagene |
| 3D3D | (1) | HYPOTHETICAL PROTEIN FROM GIARDIA LAMBLIA GL50803_14299 |
|---|
 | hypothetical, structural genomics, niaid, decode, infectious disease, seattle structural genomics center for infectious disease, ssgcid, unknown function |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PUTATIVE DIPHTHINE SYNTHASE FROM ENTAMOEBA HISTOLYTICA |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF ZINC-BOUND SUCCINYL-DIAMINOPIMELATE DESUCCINYLASE FROM HAEMOPHILUS INFLUENZAE |
|---|
 | titl: structural basis for catalysis by the mono- and dimetalated forms of the dape-encoded n-succinyl-l,l-diaminopimelic aci desuccinylase. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF GLYCINE CLEAVAGE SYSTEM PROTEIN H FROM MYCOBACTERIUM TUBERCULOSIS, USING X-RAYS FROM THE COMPACT LIGHT SOURCE. |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF CYTIDINE DEAMINASE FROM MYCOBACTERIUM TUBERCULOSIS |
|---|
 | titl: structural and functional analyses of mycobacterium tuberculosis rv3315c-encoded metal-dependent homotetrameric cytidine deaminase. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF DIHYDRODIPICOLINATE REDUCTASE FROM BARTONELLA HENSELAE AT 2.0A RESOLUTION |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF PANTOATE-BETA-ALANINE-LIGASE IN COMPLEX WITH ATP AT LOW OCCUPANCY AT 2.1 A RESOLUTION |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF MONO-ZINC FORM OF SUCCINYL-DIAMINOPIMELATE DESUCCINYLASE FROM HAEMOPHILUS INFLUENZAE |
|---|
 | titl: structural basis for catalysis by the mono- and dimetalated forms of the dape-encoded n-succinyl-l,l-diaminopimelic aci desuccinylase. |
| 3D3D | (1) | CRYO-EM STRUCTURE OF THE RABBIT VOLTAGE-GATED CALCIUM CHANNEL CAV1.1 COMPLEX AT 4.2 ANGSTROM |
|---|
 | other_details: chain b is for a subunit that is docked by a homolog crystal structure (pdb code: 1t0j) |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PEPTIDYL-PROLYL CIS-TRANS ISOMERASE FROM ENCEPHALITOZOON CUNICULI AT 1.9 A RESOLUTION |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF ENOYL-(ACYL-CARRIER-PROTEIN) REDUCTASE FROM ANAPLASMA PHAGOCYTOPHILUM AT 1.9A RESOLUTION |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF EONYL-(ACYL-CARRIER-PROTEIN) REDUCTASE FROM ANAPLASMA PHAGOCYTOPHILUM IN COMPLEX WITH NAD AT 1.9A RESOLUTION |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF A PLASMID PARTITION PROTEIN FROM BORRELIA BURGDORFERI AT 2.25A RESOLUTION, IODIDE SOAK |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF A PLASMID PARTITION PROTEIN FROM BORRELIA BURGDORFERI AT 2.25A RESOLUTION |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF THYMIDYLATE SYNTHASE 1/2 FROM ENCEPHALITOZOON CUNICULI AT 2.2 A RESOLUTION |
|---|
| 3D3D | (1) | AGAO 5-PHENOXY-2,3-PENTADIENYLAMINE COMPLEX |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF EOXYCYTIDINE TRIPHOSPHATE DEAMINASE FROM ANAPLASMA PHAGOCYTOPHILUM AT 2.1A RESOLUTION |
|---|
| 3D3D | (1) | AGAO 6-PHENYL-2,3-HEXADIENYLAMINE COMPLEX |
|---|
| 3D3D | (2) | IMP1 KH34 |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE UBIQUITIN LIKE DOMAIN OF PLXNC1 |
|---|
 | synonym: virus-encoded semaphorin protein receptor |
| 3D3D | (1) | STRUCTURE OF PHF8 IN COMPLEX WITH HISTONE H3 |
|---|
 | epigenetics, histone code, covalent histone modifications, jumonji demethylase, mental retardation, metal-binding, zinc, zinc-finger |
| 3D3D | (1) | STRUCTURE OF KIAA1718, HUMAN JUMONJI DEMETHYLASE, IN COMPLEX WITH N-OXALYLGLYCINE |
|---|
 | epigenetics, histone code, jumonji lysine demethylase, metal-binding, zinc, zinc-finger |
| 3D3D | (1) | STRUCTURE OF KIAA1718, HUMAN JUMONJI DEMETHYLASE, IN COMPLEX WITH ALPHA-KETOGLUTARATE |
|---|
 | epigenetics, histone code, jumonji lysine demethylase, metal-binding, zinc-finger, h3k4me3 binding protein, transferase |
| 3D3D | (1) | CRYSTAL STRUCTURE OF XYLULOKINASE FROM CHROMOBACTERIUM VIOLACEUM |
|---|
 | expression_system_strain: bl21(de3) coden+ril stratagene |
| 3D3D | (1) | CRYSTAL STRUCTURE ANALYSIS OF PCNA1 FROM THERMOCOCCUS KODAKARAENSIS TK0535 |
|---|
 | titl: crystal structures of two active proliferating cell nuclear antigens (pcnas) encoded by thermococcus kodakaraensis. |
| 3D3D | (1) | CRYSTAL STRUCTURE ANALYSIS OF PCNA FROM THERMOCOCCUS KODAKARAENSIS TK0582 |
|---|
 | titl: crystal structures of two active proliferating cell nuclear antigens (pcnas) encoded by thermococcus kodakaraensis. |
| 3D3D | (1) | STRUCTURE OF DROSOPHILA MELANOGASTER CARBOXYPEPTIDASE D ISOFORM 1B SHORT |
|---|
 | titl: structure-function analysis of the short splicing variant carboxypeptidase encoded by drosophila melanogaster silver. |
| 3D3D | (1) | GYMLGS SEGMENT 127-132 FROM HUMAN PRION WITH M129 |
|---|
 | titl: crystallographic studies of prion protein (prp) segments suggest how structural changes encoded by polymorphism at residue 129 modulate susceptibility to human prion disease. |
| 3D3D | (1) | GYVLGS SEGMENT 127-132 FROM HUMAN PRION WITH V129 |
|---|
 | titl: crystallographic studies of prion protein (prp) segments suggest how structural changes encoded by polymorphism at residue 129 modulate susceptibility to human prion disease. |
| 3D3D | (1) | MOLECULAR MECHANISM OF GUIDANCE CUE RECOGNITION |
|---|
 | synonym: virus-encoded semaphorin protein receptor |
| 3D3D | (1) | MOLECULAR MECHANISM OF GUIDANCE CUE RECOGNITION |
|---|
 | synonym: virus-encoded semaphorin protein receptor |
| 3D3D | (1) | THE STRUCTURE OF CA2+ SENSOR (CASE-16) |
|---|
| 3D3D | (1) | THE STRUCTURE OF CA2+ SENSOR (CASE-12) |
|---|
| 3D3D | (1) | THE SECRET DOMAIN FROM ECTROMELIA VIRUS |
|---|
 | titl: structural basis of chemokine sequestration by crmd, a poxvirus-encoded tumor necrosis factor receptor |
| 3D3D | (1) | THE SECRET DOMAIN IN COMPLEX WITH CX3CL1 |
|---|
 | titl: structural basis of chemokine sequestration by crmd, a poxvirus-encoded tumor necrosis factor receptor |
| 3D3D | (1) | I-SCEI COMPLEXED WITH C/G+4 DNA SUBSTRATE |
|---|
 | molecule: intron encoded endonuclease i-scei |
| 3D3D | (1) | I-SCEI MUTANT (K86R/G100T)COMPLEXED WITH C/G+4 DNA SUBSTRATE |
|---|
 | molecule: intron encoded endonuclease i-scei |
| 3D3D | (1) | MALTOSE-BOUND MALTOSE SENSOR ENGINEERED BY INSERTION OF CIRCULARLY PERMUTED GREEN FLUORESCENT PROTEIN INTO E. COLI MALTOSE BINDING PROTEIN AT POSITION 175 |
|---|
 | titl: a genetically encoded, high-signal-to-noise maltose sensor. |
| 3D3D | (1) | MALTOSE-BOUND MALTOSE SENSOR ENGINEERED BY INSERTION OF CIRCULARLY PERMUTED GREEN FLUORESCENT PROTEIN INTO E. COLI MALTOSE BINDING PROTEIN AT POSITION 311 |
|---|
 | titl: a genetically encoded, high-signal-to-noise maltose sensor. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE 30S RIBOSOMAL SUBUNIT FROM A KSGA MUTANT OF THERMUS THERMOPHILUS (HB8) |
|---|
 | 30s ribosomal subunit, ksga knock-out, post transcriptional rrna modification, antibiotic resistance, decoding, decoding of genetic code, trna, mrna, ribosome |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A PUTATIVE GLYCOSYLTRANSFERASE FROM PARAMECIUM BURSARIA CHLORELLA VIRUS NY2A |
|---|
 | titl: crystal structure of a virus-encoded putative glycosyltransferase |
| 3D3D | (2) | CRYSTAL STRUCTURE OF A VIRUS ENCODED GLYCOSYLTRANSFERASE IN COMPLEX WITH GDP-MANNOSE |
|---|
 | titl: crystal structure of a virus-encoded putative glycosyltransferase. | |
| crystal structure of a virus encoded glycosyltransferase in complex with gdp-mannose |
| 3D3D | (1) | STRUCTURE OF GFCC (YMCB), PROTEIN ENCODED BY THE E. COLI GROUP 4 CAPSULE OPERON |
|---|
 | structure of gfcc (ymcb), protein encoded by the e. coli group 4 capsule operon |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH IBET INHIBITOR |
|---|
 | auth: e.nicodeme,k.l.jeffrey,u.schaefer,s.beinke,s.dewell,c.chung r.chandwani,i.marazzi,p.wilson,h.coste,j.white,j.kirilovsky c.m.rice,j.m.lora,r.k.prinjha,k.lee,a.tarakhovsky |
| 3D3D | (1) | HCV NS5B WITH A BOUND QUINOLONE INHIBITOR |
|---|
 | molecule: hcv encoded nonstructural 5b protein |
| 3D3D | (1) | TRYPTOPHANYL-TRNA SYNTHETASE VAL144PRO MUTANT FROM B. SUBTILIS |
|---|
 | titl: importance of single molecular determinants in the fidelity of expanded genetic codes. |
| 3D3D | (1) | STRUCTURE OF ESPG PROTEIN |
|---|
 | molecule: lee-encoded effector espg |
| 3D3D | (2) | STRUCTURE OF YGEW ENCODED PROTEIN FROM E. COLI |
|---|
 | structure of ygew encoded protein from e. coli | |
| titl: the ygew encoded protein from escherichia coli is a knotted ancestral catabolic transcarbamylase. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE SERYL-TRNA SYNTHETASE FROM CANDIDA ALBICANS |
|---|
 | genetic code ambiguity, trna, class-ii aminoacyl-trna synthetase family, type-1 seryl-trna synthetase subfamily, trna aminoacylation, serine, ligase |
| 3D3D | (1) | HK97 PROHEAD I ENCAPSIDATING INACTIVE VIRALLY ENCODED PROTEASE |
|---|
 | hk97 prohead i encapsidating inactive virally encoded protease |
| 3D3D | (1) | STRUCTURE OF N15 CRO COMPLEXED WITH CONSENSUS OPERATOR DNA |
|---|
 | titl: mysteries of an evolutionary code: the structure of n15 cro complexed with operator dna |
| 3D3D | (1) | CRYSTAL STRUCTURE OF MAMMALIAN CAPPING ENZYME (MCE1) AND POL II CTD COMPLEX |
|---|
 | titl: structural insights to how mammalian capping enzyme reads the ctd code. |
| 3D3D | (1) | THE CRYSTAL STRUCTURE OF A HYDROLASE FROM PSEUDOMONAS SYRINGAE |
|---|
 | expression_system_strain: bl21(de3)-coden+ril(p)-stratagene |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE PASSENGER DOMAIN OF THE E. COLI AUTOTRANSPORTER ESPP |
|---|
 | synonym: secreted autotransporter protein espp, extracellular serine protease plasmid-encoded espp |
| 3D3D | (1) | HUMAN PIN1 BOUND TO CIS PEPTIDOMIMETIC INHIBITOR |
|---|
 | titl: structural and kinetic analysis of prolyl-isomerization/phosphorylation cross-talk in the ctd code. |
| 3D3D | (1) | HUMAN PIN1 BOUND TO TRANS PEPTIDOMIMETIC INHIBITOR |
|---|
 | titl: structural and kinetic analysis of prolyl-isomerization/phosphorylation cross-talk in the ctd code. |
| 3D3D | (3) | CRYSTAL STRUCTURE OF THE GENETICALLY ENCODED CALCIUM INDICATOR RCAMP |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE ENGINEERED FLUORESCENT PROTEIN MRUBY, CRYSTAL FORM 1, PH 4.5 |
|---|
 | titl: genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE ENGINEERED FLUORESCENT PROTEIN MRUBY, CRYSTAL FORM 1, PH 8.5 |
|---|
 | titl: genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE ENGINEERED FLUORESCENT PROTEIN MRUBY, CRYSTAL FORM 2 |
|---|
 | titl: genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. |
| 3D3D | (2) | STRUCTURES OF ALKALOID BIOSYNTHETIC GLUCOSIDASES DECODE SUBSTRATE SPECIFICITY |
|---|
 | titl: structures of alkaloid biosynthetic glucosidases decode substrate specificity. | |
| structures of alkaloid biosynthetic glucosidases decode substrate specificity |
| 3D3D | (2) | STRUCTURES OF ALKALOID BIOSYNTHETIC GLUCOSIDASES DECODE SUBSTRATE SPECIFICITY |
|---|
 | structures of alkaloid biosynthetic glucosidases decode substrate specificity | |
| titl: structures of alkaloid biosynthetic glucosidases decode substrate specificity. |
| 3D3D | (2) | STRUCTURES OF ALKALOID BIOSYNTHETIC GLUCOSIDASES DECODE SUBSTRATE SPECIFICITY |
|---|
 | titl: structures of alkaloid biosynthetic glucosidases decode substrate specificity. | |
| structures of alkaloid biosynthetic glucosidases decode substrate specificity |
| 3D3D | (2) | CRYSTAL STRUCTURE OF A TRP-LESS GREEN FLUORESCENT PROTEIN TRANSLATED BY THE UNIVERSAL GENETIC CODE |
|---|
| 3D3D | (2) | CRYSTAL STRUCTURE OF A TRP-LESS GREEN FLUORESCENT PROTEIN TRANSLATED BY THE SIMPLIFIED GENETIC CODE |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE CPSRP54 TAIL BOUND TO CPSRP43 |
|---|
 | titl: chromodomains read the arginine code of post-translational targeting. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF ANTI-HIV-1 V3 FAB 2558 IN COMPLEX WITH MN PEPTIDE |
|---|
 | titl: human anti-v3 hiv-1 monoclonal antibodies encoded by the vh5-51/vl lambda genes define a conserved antigenic structure. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF ANTI-HIV-1 V3 FAB 4025 IN COMPLEX WITH CON A PEPTIDE |
|---|
 | titl: human anti-v3 hiv-1 monoclonal antibodies encoded by the vh5-51/vl lambda genes define a conserved antigenic structure. |
| 3D3D | (1) | EXPANDING LAGALIDADG ENDONUCLEASE SCAFFOLD DIVERSITY BY RAPIDLY SURVEYING EVOLUTIONARY SEQUENCE SPACE |
|---|
 | molecule: intron-encoded dna endonuclease i-hjemi |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A PROBABLE PEPTIDE DEFORMYLASE FROM SYNECHOCOCCUS PHAGE S-SSM7 |
|---|
 | titl: structure and function of a cyanophage-encoded peptide deformylase. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A PROBABLE PEPTIDE DEFORMYLASE FROM STRUCYNECHOCOCCUS PHAGE S-SSM7 IN COMPLEX WITH ACTINONIN |
|---|
 | titl: structure and function of a cyanophage-encoded peptide deformylase. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF CLOSTRIDIUM BOTULINUM PHAGE C-ST TUBZ |
|---|
 | titl: tubulin homolog tubz in a phage-encoded partition system. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF MNMC2 FROM AQUIFEX AEOLICUS |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF MONOMERIC PHOTOSENSITIZING FLUORESCENT PROTEIN, SUPERNOVA |
|---|
 | gfp fold, fluorescent protein, genetically encoded photosensitizer, phototoxicity |
| 3D3D | (1) | THE CRYSTAL STRUCTURE OF NATIVE GLYCOSIDIC HYDROLASE |
|---|
 | titl: structural analyses and yeast production of the beta-1,3-1,4-glucanase catalytic module encoded by the licb gene of clostridium thermocellum. |
| 3D3D | (1) | STRUCTURE OF BSUDG-P56 COMPLEX |
|---|
 | synonym: left end of bacteriophage phi-29 coding for 15 potential proteins among these are the terminal protein and the proteins encoded by the genes 1,2 (sus), 3, and (probably) 4 |
| 3D3D | (1) | STRUCTURE OF TETRACYCLINE REPRESSOR IN COMPLEX WITH ANTIINDUCER PEPTIDE-TAP1 |
|---|
 | titl: an exclusive alpha/beta code directs allostery in tetr-peptide complexes. |
| 3D3D | (1) | STRUCTURE OF TETRACYCLINE REPRESSOR IN COMPLEX WITH ANTIINDUCER PEPTIDE-TAP2 |
|---|
 | titl: an exclusive alpha/beta code directs allostery in tetr-peptide complexes. |
| 3D3D | (1) | STRUCTURE OF TETRACYCLINE REPRESSOR IN COMPLEX WITH INDUCER PEPTIDE-TIP3 |
|---|
 | titl: an exclusive alpha/beta code directs allostery in tetr-peptide complexes. |
| 3D3D | (1) | STRUCTURE OF TETRACYCLINE REPRESSOR IN COMPLEX WITH INDUCER PEPTIDE-TIP2 |
|---|
 | titl: an exclusive alpha/beta code directs allostery in tetr-peptide complexes. |
| 3D3D | (1) | 5'-D(*AP*CP*CP*GP*AP*CP*GP*TP*CP*GP*GP*T)-3' |
|---|
 | titl: structural code for dna recognition revealed in crystal structures of papillomavirus e2-dna targets. |
| 3D3D | (1) | 5'-D(*AP*CP*CP*GP*AP*CP*GP*TP*CP*GP*GP*T)-3' |
|---|
 | titl: structural code for dna recognition revealed in crystal structures of papillomavirus e2-dna targets. |
| 3D3D | (1) | 5'-D(*AP*CP*CP*GP*GP*TP*AP*CP*CP*GP*GP*T)-3' |
|---|
 | titl: structural code for dna recognition revealed in crystal structures of papillomavirus e2-dna targets. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF RAUCAFFRICINE GLUCOSIDASE FROM AJMALINE BIOSYNTHESIS PATHWAY |
|---|
 | titl: structures of alkaloid biosynthetic glucosidases decode substrate specificity. |
| 3D3D | (1) | N-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH TBUTYL-PHENYL-AMINO-DIMETHYL-OXAZOLYL-QUINOLINE-CARBOXYLIC ACID |
|---|
| 3D3D | (1) | N-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH IBET-151 |
|---|
| 3D3D | (2) | A MEGAVIRIDAE ORFAN GENE ENCODES A NEW NUCLEOTIDYL TRANSFERASE |
|---|
 | titl: a megaviridae orphan gene encodes a new nucleotidyl transferase | |
| a megaviridae orfan gene encodes a new nucleotidyl transferase |
| 3D3D | (2) | A MEGAVIRIDAE ORFAN GENE ENCODE A NEW NUCLEOTIDYL TRANSFERASE |
|---|
 | titl: a megaviridae orphan gene encodes a new nucleotidyl transferase | |
| a megaviridae orfan gene encode a new nucleotidyl transferase |
| 3D3D | (1) | THE CRYSTAL STRUCTURE OF CHRYSONILIA SITOPHILA ENDO-BETA-D-1,4-MANNANASE |
|---|
| 3D3D | (1) | NEW TYPE VI-SECRETED TOXINS AND SELF-RESISTANCE PROTEINS IN SERRATIA MARCESCENS |
|---|
 | titl: new secreted toxins and immunity proteins encoded within the type vi secretion system gene cluster of serratia marcescens |
| 3D3D | (1) | CRYSTAL STRUCTURE RAP2B (SMA2266) FROM SERRATIA MARCESCENS |
|---|
 | titl: new secreted toxins and immunity proteins encoded within th type vi secretion system gene cluster of serratia marcescens. |
| 3D3D | (1) | BACTERIOPHAGE T5 HOMOLOG OF THE EUKARYOTIC TRANSCRIPTION COACTIVATOR PC4 IMPLICATED IN RECOMBINATION-DEPENDENT DNA REPLICATION |
|---|
 | titl: bacteriophage t5 encodes a homolog of the eukaryotic transcription coactivator pc4 implicated in recombination- dependent dna replication. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE HUMAN EPHA4 ECTODOMAIN |
|---|
 | titl: structurally encoded intraclass differences in epha clusters drive distinct cell responses |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE HUMAN EPHA4 ECTODOMAIN IN COMPLEX WITH HUMAN EPHRIN A5 (AMINE-METHYLATED SAMPLE) |
|---|
 | titl: structurally encoded intraclass differences in epha clusters drive distinct cell responses |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE HUMAN EPHA4 ECTODOMAIN IN COMPLEX WITH HUMAN EPHRIN A5 |
|---|
 | titl: structurally encoded intraclass differences in epha clusters drive distinct cell responses |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE HUMAN EPHA4 ECTODOMAIN IN COMPLEX WITH HUMAN EPHRINB3 |
|---|
 | titl: structurally encoded intraclass differences in epha clusters drive distinct cell responses |
| 3D3D | (1) | THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH 3,5 DIMETHYLISOXAXOLE LIGAND |
|---|
 | auth: o.mirguet,y.lamotte,c.chung,p.bamborough,d.delannee, a.bouillot,f.gellibert,g.krysa,a.lewis,j.witherington, p.huet,y.dudit,l.trottet,e.nicodeme |
| 3D3D | (1) | THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH 3,5 DIMETHYLISOXAXOLE LIGAND |
|---|
 | auth: o.mirguet,y.lamotte,c.chung,p.bamborough,d.delannee, a.bouillot,f.gellibert,g.krysa,a.lewis,j.witherington, p.huet,y.dudit,l.trottet,e.nicodeme |
| 3D3D | (1) | THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH 3,5 DIMETHYLISOXAXOLE LIGAND |
|---|
 | auth: o.mirguet,y.lamotte,c.chung,p.bamborough,d.delannee, a.bouillot,f.gellibert,g.krysa,a.lewis,j.witherington, p.huet,y.dudit,l.trottet,e.nicodeme |
| 3D3D | (1) | THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH 3,5 DIMETHYLISOXAXOLE LIGAND |
|---|
 | auth: o.mirguet,y.lamotte,c.chung,p.bamborough,d.delannee, a.bouillot,f.gellibert,g.krysa,a.lewis,j.witherington, p.huet,y.dudit,l.trottet,e.nicodeme |
| 3D3D | (1) | COILED COIL DOMAIN OF THE ZFL2-1 ORF1 PROTEIN FROM THE ZEBRAFISH ZFL2-1 RETROTRANSPOSON |
|---|
 | molecule: orf1-encoded protein |
| 3D3D | (1) | ESTERASE DOMAIN OF THE ZFL2-1 ORF1 PROTEIN FROM THE ZEBRAFISH ZFL2-1 RETROTRANSPOSON |
|---|
 | molecule: orf1-encoded protein |
| 3D3D | (1) | DISCOVERY OF EPIGENETIC REGULATOR I-BET762: LEAD OPTIMIZATION TO AFFORD A CLINICAL CANDIDATE INHIBITOR OF THE BET BROMODOMAINS |
|---|
| 3D3D | (1) | DISCOVERY OF EPIGENETIC REGULATOR I-BET762: LEAD OPTIMIZATION TO AFFORD A CLINICAL CANDIDATE INHIBITOR OF THE BET BROMODOMAINS |
|---|
| 3D3D | (3) | ENCODED LIBRARY TECHNOLOGY AS A SOURCE OF HITS FOR THE DISCOVERY AND LEAD OPTIMIZATION OF A POTENT AND SELECTIVE CLASS OF BACTERICIDAL DIRECT INHIBITORS OF MYCOBACTERIUM TUBERCULOSIS INHA |
|---|
| 3D3D | (1) | REGULATION OF THE MAMMALIAN ELONGATION CYCLE BY 40S SUBUNIT ROLLING: A EUKARYOTIC-SPECIFIC RIBOSOME REARRANGEMENT |
|---|
 | other_details: for modeling mrna (chain 1, pdb id code 3i8g) was used |
| 3D3D | (1) | REGULATION OF THE MAMMALIAN ELONGATION CYCLE BY 40S SUBUNIT ROLLING: A EUKARYOTIC-SPECIFIC RIBOSOME REARRANGEMENT |
|---|
 | other_details: for modeling mrna (chain 1, pdb id code 3i8g) was used |
| 3D3D | (2) | DISCOVERY OF GLYCOMIMETIC LIGANDS VIA GENETICALLY-ENCODED LIBRARY OF PHAGE DISPLAYING MANNOSE-PEPTIDES |
|---|
| 3D3D | (2) | STRUCTURAL AND FUNCTIONAL STUDIES OF THE TRANS-ENCODED HLA-DQ2.3 (DQA1*03:01/DQB1*02:01) MOLECULE |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE APO 30S RIBOSOMAL SUBUNIT FROM THERMUS THERMOPHILUS (HB8) |
|---|
 | titl: a structural basis for streptomycin-induced misreading of the genetic code. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE THERMUS THERMOPHILUS (HB8) 30S RIBOSOMAL SUBUNIT WITH MULTIPLE COPIES OF PAROMOMYCIN MOLECULES BOUND |
|---|
 | titl: a structural basis for streptomycin-induced misreading of the genetic code. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE THERMUS THERMOPHILUS (HB8) 30S RIBOSOMAL SUBUNIT WITH STREPTOMYCIN BOUND |
|---|
 | titl: a structural basis for streptomycin-induced misreading of the genetic code. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE THERMUS THERMOPHILUS (HB8) 30S RIBOSOMAL SUBUNIT WITH CODON, COGNATE TRANSFER RNA ANTICODON STEM-LOOP AND MULTIPLE COPIES OF PAROMOMYCIN MOLECULES BOUND |
|---|
 | titl: a structural basis for streptomycin-induced misreading of the genetic code. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE THERMUS THERMOPHILUS (HB8) 30S RIBOSOMAL SUBUNIT WITH CODON, CRYSTALLOGRAPHICALLY DISORDERED COGNATE TRANSFER RNA ANTICODON STEM-LOOP AND STREPTOMYCIN BOUND |
|---|
 | titl: a structural basis for streptomycin-induced misreading of the genetic code. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE THERMUS THERMOPHILUS (HB8) 30S RIBOSOMAL SUBUNIT WITH CODON, NEAR-COGNATE TRANSFER RNA ANTICODON STEM-LOOP MISMATCHED AT THE FIRST CODON POSITION AND STREPTOMYCIN BOUND |
|---|
 | titl: a structural basis for streptomycin-induced misreading of the genetic code. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE THERMUS THERMOPHILUS (HB8) 30S RIBOSOMAL SUBUNIT WITH CODON, CRYSTALLOGRAPHICALLY DISORDERED NEAR-COGNATE TRANSFER RNA ANTICODON STEM-LOOP MISMATCHED AT THE SECOND CODON POSITION, AND STREPTOMYCIN BOUND |
|---|
 | titl: a structural basis for streptomycin-induced misreading of the genetic code. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A PEPTIDE DEFORMYLASE FROM SYNECHOCOCCUS ELONGATUS |
|---|
 | titl: structure and function of a cyanophage-encoded peptide deformylase. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A PEPTIDE DEFORMYLASE FROM SYNECHOCOCCUS ELONGATUS IN COMPLEX WITH ACTINONIN |
|---|
 | titl: structure and function of a cyanophage-encoded peptide deformylase. |
| 3D3D | (1) | NOVEL AND SELECTIVE PAN-PIM KINASE INHIBITOR |
|---|
 | auth: l.a.dakin,m.h.block,h.chen,e.code,j.e.dowling,x.feng, a.d.ferguson,i.green,a.w.hird,t.howard,e.k.keeton,m.l.lamb, p.d.lyne,h.pollard,j.read,a.j.wu,t.zhang,x.zheng |
| 3D3D | (1) | CRYSTAL STRUCTURE OF ORFX IN COMPLEX WITH S-ADENOSYLMETHIONINE |
|---|
 | titl: characterization of the staphylococcus aureus rrna methyltransferase encoded by orfx, the gene containing the staphylococcal chromosome cassette mec (sccmec) insertion site. |
| 3D3D | (1) | THE STRUCTURE OF COWPOX VIRUS CPXV018 (OMCP) |
|---|
 | titl: crystal structure of the cowpox virus-encoded nkg2d ligand omcp. |
| 3D3D | (1) | PYLC IN COMPLEX WITH L-LYSINE |
|---|
 | titl: biosynthesis of the 22nd genetically encoded amino acid pyrrolysine: structure and reaction mechanism of pylc at 1.5a resolution. |
| 3D3D | (1) | PYLC IN COMPLEX WITH L-LYSINE-NE-D-ORNITHINE (COCRYSTALLIZED WITH L-LYSINE-NE-D-ORNITHINE) |
|---|
 | titl: biosynthesis of the 22nd genetically encoded amino acid pyrrolysine: structure and reaction mechanism of pylc at 1.5a resolution. |
| 3D3D | (1) | PYLC IN COMPLEX WITH D-ORNITHINE AND AMPPNP |
|---|
 | titl: biosynthesis of the 22nd genetically encoded amino acid pyrrolysine: structure and reaction mechanism of pylc at 1.5a resolution. |
| 3D3D | (1) | PYLC IN COMPLEX WITH PHOSPHORYLATED D-ORNITHINE |
|---|
 | titl: biosynthesis of the 22nd genetically encoded amino acid pyrrolysine: structure and reaction mechanism of pylc at 1.5a resolution. |
| 3D3D | (1) | PYLC IN COMPLEX WITH L-LYSINE-NE-D-ORNITHINE (COCRYSTALLIZED WITH L-LYSINE AND D-ORNITHINE) |
|---|
 | titl: biosynthesis of the 22nd genetically encoded amino acid pyrrolysine: structure and reaction mechanism of pylc at 1.5a resolution. |
| 3D3D | (1) | SEMET-LABELED PYLC (REMOTE) |
|---|
 | titl: biosynthesis of the 22nd genetically encoded amino acid pyrrolysine: structure and reaction mechanism of pylc at 1.5a resolution. |
| 3D3D | (1) | ALLOSTERIC PEPTIDIC INHIBITOR OF HUMAN THYMIDYLATE SYNTHASE THAT STABILIZES INACTIVE CONFORMATION OF THE ENZYME. |
|---|
 | titl: alanine mutants of the interface residues of human thymidylate synthase decode key features of the binding mod of allosteric anticancer peptides. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF RNASE Z IN COMPLEX WITH PRECURSOR TRNA(THR) |
|---|
 | other_details: b.subtilis trnathr(encoded by the trn1-thr gene) was synthetised as a precursor with 2 nt 3'-extension. the nt 73-76 are 2'-o-methylated. |
| 3D3D | (1) | STRUCTURE OF MUTS139F P73 DNA BINDING DOMAIN COMPLEXED WITH 20BP DNA RESPONSE ELEMENT |
|---|
 | titl: transactivation specificity is conserved among p53 family proteins and depends on a response element sequence code. |
| 3D3D | (2) | CRYSTAL STRUCTURE OF THE VIRALLY ENCODED ANTIFUNGAL PROTEIN, KP6, HETERODIMER |
|---|
 | titl: the atomic structure of the virally encoded antifungal protein, kp6. | |
| crystal structure of the virally encoded antifungal protein, kp6, heterodimer |
| 3D3D | (1) | CRYSTAL STRUCTURE OF F2YRS |
|---|
 | titl: a genetically encoded 19f nmr probe for tyrosine phosphorylation. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF F2YRS COMPLEXED WITH F2Y |
|---|
 | titl: a genetically encoded 19f nmr probe for tyrosine phosphorylation. |
| 3D3D | (1) | R2-LIKE LIGAND-BINDING OXIDASE WITH AEROBICALLY RECONSTITUTED METAL COFACTOR |
|---|
 | titl: direct observation of structurally encoded metal discrimination and ether bond formation in a heterodinuclea metalloprotein |
| 3D3D | (1) | R2-LIKE LIGAND-BINDING OXIDASE WITH ANAEROBICALLY RECONSTITUTED METAL COFACTOR |
|---|
 | titl: direct observation of structurally encoded metal discrimination and ether bond formation in a heterodinuclea metalloprotein |
| 3D3D | (1) | R2-LIKE LIGAND-BINDING OXIDASE WITHOUT METAL COFACTOR |
|---|
 | titl: direct observation of structurally encoded metal discrimination and ether bond formation in a heterodinuclea metalloprotein |
| 3D3D | (2) | CRYSTAL STRUCTURE OF THE GENETICALLY ENCODED CALCIUM INDICATOR RGECO1 |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF NUCLEOCAPSID PROTEIN ENCODED BY THE PROTOTYPIC MEMBER OF ORTHOBUNYAVIRUS |
|---|
 | crystal structure of nucleocapsid protein encoded by the prototypic member of orthobunyavirus |
| 3D3D | (1) | DARK-STATE STRUCTURE OF SFGFP CONTAINING THE UNNATURAL AMINO ACID P-AZIDO-PHENYLALANINE AT RESIDUE 66 |
|---|
 | titl: different photochemical events of a genetically encoded phenyl azide define and modulate gfp fluorescence. |
| 3D3D | (2) | DIFFERENT PHOTOCHEMICAL EVENTS OF A GENETICALLY ENCODED ARYL AZIDE DEFINE AND MODULATE GFP FLUORESCENCE |
|---|
| 3D3D | (1) | IRRADIATED-STATE STRUCTURE OF SFGFP CONTAINING THE UNNATURAL AMINO ACID P-AZIDO-PHENYLALANINE AT RESIDUE 145 |
|---|
 | titl: different photochemical events of a genetically encoded phenyl azide define and modulate gfp fluorescence. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF AN INACTIVE MUTANT OF MMP-9 CATALYTIC DOMAIN IN COMPLEX WITH A FLUOROGENIC SYNTHETIC PEPTIDIC SUBSTRATE WITH A FLUORINE ATOM. |
|---|
 | other_details: fluorogenic synthetic l-peptide substrate with lower catalytic efficiency with mmp-9 than the iodine containing substrate pdb code 4jij. |
| 3D3D | (1) | DISTAL STEM I REGION FROM G. KAUSTOPHILUS GLYQS T BOX RNA |
|---|
 | titl: t box rna decodes both the information content and geometry of trna to affect gene expression. |
| 3D3D | (1) | STRUCTURAL AND FUNCTIONAL CHARACTERIZATION OF PSEUDOMONAS AERUGINOSA ALGX |
|---|
 | auth: l.m.riley,j.t.weadge,p.baker,h.robinson,j.d.codee, p.a.tipton,d.e.ohman,p.l.howell |
| 3D3D | (1) | X-RAY STRUCTURE OF ENDOLYSIN FROM CLOSTRIDIUM PERFRINGENS PHAGE PHISM101 |
|---|
 | titl: x-ray structure of a novel endolysin encoded by episomal phage phism101 of clostridium perfringens |
| 3D3D | (1) | X-RAY STRUCTURE OF CATALYTIC DOMAIN OF ENDOLYSIN FROM CLOSTRIDIUM PERFRINGENS PHAGE PHISM101 |
|---|
 | titl: x-ray structure of a novel endolysin encoded by episomal phage phism101 of clostridium perfringens |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE I-SMAMI LAGLIDADG HOMING ENDONUCLEASE BOUND TO CLEAVED DNA |
|---|
 | gene: cytochrome c oxidase subunit 1, mitochondrial introne encoded, smac_12671 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF APO-BRD4(1) |
|---|
 | titl: 4-acyl pyrroles: mimicking acetylated lysines in histone code reading. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF BRD4(1) BOUND TO COLCHICEINE |
|---|
 | titl: 4-acyl pyrroles: mimicking acetylated lysines in histone code reading. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF BRD4(1) BOUND TO INHIBITOR XD14 |
|---|
 | titl: 4-acyl pyrroles: mimicking acetylated lysines in histone code reading. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF BRD4(1) BOUND TO COLCHICINE |
|---|
 | titl: 4-acyl pyrroles: mimicking acetylated lysines in histone code reading. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF BRD4(1) BOUND TO INHIBITOR XD46 |
|---|
 | titl: 4-acyl pyrroles: mimicking acetylated lysines in histone code reading. |
| 3D3D | (1) | THE CRYSTAL STRUCTURE OF STX2 AND A DISACCHARIDE LIGAND |
|---|
 | synonym: shiga toxin 2 subunit a, shiga-like toxin ii a subunit encoded by bacteriophage bp-933w |
| 3D3D | (1) | CRYSTAL STRUCTURE OF SALMONELLA EFFECTOR PROTEIN GTGE |
|---|
 | molecule: bacteriophage encoded virulence factor |
| 3D3D | (1) | DISULFIDE ISOMERASE (DSBP) FROM MULTIDRUG RESISTANCE INCA/C TRANSFERABLE PLASMID IN OXIDIZED STATE (P212121 SPACE GROUP) |
|---|
 | titl: the multidrug resistance inca/c transferable plasmid encode a novel domain-swapped dimeric protein-disulfide isomerase. |
| 3D3D | (1) | DISULFIDE ISOMERASE FROM MULTIDRUG RESISTANCE INCA/C CONJUGATIVE PLASMID IN REDUCED STATE |
|---|
 | titl: the multidrug resistance inca/c transferable plasmid encode a novel domain-swapped dimeric protein-disulfide isomerase. |
| 3D3D | (1) | DISULFIDE ISOMERASE FROM MULTIDRUG RESISTANCE INCA/C RELATED INTEGRATIVE AND CONJUGATIVE ELEMENTS IN OXIDIZED STATE (P21 SPACE GROUP) |
|---|
 | titl: the multidrug resistance inca/c transferable plasmid encode a novel domain-swapped dimeric protein-disulfide isomerase. |
| 3D3D | (1) | 1.45 A RESOLUTION CRYSTAL STRUCTURE OF PROTEIN PHOSPHATASE 1 |
|---|
 | titl: understanding the antagonism of retinoblastoma protein dephosphorylation by pnuts provides insights into the pp1 regulatory code. |
| 3D3D | (1) | STRUCTURE OF A SECOND NUCLEAR PP1 HOLOENZYME, CRYSTAL FORM 1 |
|---|
 | titl: understanding the antagonism of retinoblastoma protein dephosphorylation by pnuts provides insights into the pp1 regulatory code. |
| 3D3D | (1) | STRUCTURE OF A SECOND NUCLEAR PP1 HOLOENZYME, CRYSTAL FORM 2 |
|---|
 | titl: understanding the antagonism of retinoblastoma protein dephosphorylation by pnuts provides insights into the pp1 regulatory code. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF FOSB FROM STAPHYLOCOCCUS AUREUS WITH ZN AND SULFATE AT 1.42 ANGSTROM RESOLUTION - SAD PHASING |
|---|
 | titl: structure and function of the genomically encoded fosfomyci resistance enzyme, fosb, from staphylococcus aureus. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF FOSB FROM STAPHYLOCOCCUS AUREUS WITH ZN AND SULFATE AT 1.15 ANGSTROM RESOLUTION |
|---|
 | titl: structure and function of the genomically encoded fosfomyci resistance enzyme, fosb, from staphylococcus aureus. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF FOSB FROM STAPHYLOCOCCUS AUREUS WITH BS-CYS9 DISULFIDE AT 1.62 ANGSTROM RESOLUTION |
|---|
 | titl: structure and function of the genomically encoded fosfomyci resistance enzyme, fosb, from staphylococcus aureus. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF FOSB FROM STAPHYLOCOCCUS AUREUS AT 1.80 ANGSTROM RESOLUTION WITH L-CYSTEINE-CYS9 DISULFIDE |
|---|
 | titl: structure and function of the genomically encoded fosfomyci resistance enzyme, fosb, from staphylococcus aureus. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF FOSB FROM STAPHYLOCOCCUS AUREUS AT 1.89 ANGSTROM RESOLUTION - APO STRUCTURE |
|---|
 | titl: structure and function of the genomically encoded fosfomyci resistance enzyme, fosb, from staphylococcus aureus. |
| 3D3D | (1) | D-AMINOACYL-TRNA DEACYLASE (DTD) FROM PLASMODIUM FALCIPARUM IN COMPLEX WITH D-TYROSYL-3'-AMINOADENOSINE AT 1.86 ANGSTROM RESOLUTION |
|---|
 | titl: mechanism of chiral proofreading during translation of the genetic code. |
| 3D3D | (1) | D-AMINOACYL-TRNA DEACYLASE (DTD) FROM PLASMODIUM FALCIPARUM IN COMPLEX WITH D-TYROSYL-3'-AMINOADENOSINE AT 2.20 ANGSTROM RESOLUTION |
|---|
 | titl: mechanism of chiral proofreading during translation of the genetic code. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE LACTATE DEHYDROGENASE FROM CRYPTOSPORIDIUM PARVUM COMPLEXED WITH COFACTOR (B-NICOTINAMIDE ADENINE DINUCLEOTIDE) AND INHIBITOR (OXAMIC ACID) |
|---|
 | molecule: lactate dehydrogenase, adjacent gene encodes predicted malate dehydrogenase |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE LACTATE DEHYDROGENASE FROM CRYPTOSPORIDIUM PARVUM COMPLEXED WITH SUBSTRATE (PYRUVIC ACID) AND COFACTOR ANALOG (3-ACETYLPYRIDINE ADENINE DINUCLEOTIDE) |
|---|
 | molecule: lactate dehydrogenase, adjacent gene encodes predicted malate dehydrogenase |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE LACTATE DEHYDROGENASE FROM CRYPTOSPORIDIUM PARVUM COMPLEXED WITH SUBSTRATE (L-LACTIC ACID) AND COFACTOR (B-NICOTINAMIDE ADENINE DINUCLEOTIDE) |
|---|
 | molecule: lactate dehydrogenase, adjacent gene encodes predicted malate dehydrogenase |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE LACTATE DEHYDROGENASE FROM CRYPTOSPORIDIUM PARVUM COMPLEXED WITH SUBSTRATE (PYRUVIC ACID) AND COFACTOR (B-NICOTINAMIDE ADENINE DINUCLEOTIDE) |
|---|
 | molecule: lactate dehydrogenase, adjacent gene encodes predicted malate dehydrogenase |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE LACTATE DEHYDROGENASE FROM CRYPTOSPORIDIUM PARVUM |
|---|
 | molecule: lactate dehydrogenase, adjacent gene encodes predicted malate dehydrogenase |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE PERIPLASMIC ALGINATE EPIMERASE ALGG |
|---|
 | auth: f.wolfram,e.n.kitova,h.robinson,m.t.walvoort,j.d.codee, j.s.klassen,p.l.howell |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE PERIPLASMIC ALGINATE EPIMERASE ALGG D317A MUTANT |
|---|
 | auth: f.wolfram,e.n.kitova,h.robinson,m.t.walvoort,j.d.codee, j.s.klassen,p.l.howell |
| 3D3D | (1) | STRUCTURE OF AN AMYLOID FORMING PEPTIDE VQIVYK FROM THE SECOND REPEAT REGION OF TAU (ALTERNATE POLYMORPH) |
|---|
 | titl: molecular mechanisms for protein-encoded inheritance. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF MONO-ZINC FORM OF SUCCINYL DIAMINOPIMELATE DESUCCINYLASE FROM NEISSERIA MENINGITIDIS MC58 |
|---|
 | titl: inhibition of the dape-encoded n-succinyl-l,l-diaminopimeli acid desuccinylase from neisseria meningitidis by l-captopril. |
| 3D3D | (3) | THE CRYSTAL STRUCTURE OF NON-LEE ENCODED TYPE III EFFECTOR C FROM CITROBACTER RODENTIUM |
|---|
| 3D3D | (1) | O-ACETYLTRANSFERASE DOMAIN OF PSEUDOMONAS PUTIDA ALGJ |
|---|
 | auth: p.baker,t.ricer,p.j.moynihan,e.n.kitova,m.t.walvoort, d.j.little,j.c.whitney,k.dawson,j.t.weadge,h.robinson, d.e.ohman,j.d.codee,j.s.klassen,a.j.clarke,p.l.howell |
| 3D3D | (1) | 4E10 GERMLINE ENCODED PRECURSOR NO.7 IN COMPLEX WITH EPITOPE SCAFFOLD T117 |
|---|
 | 4e10 germline encoded precursor no.7 in complex with epitope scaffold t117 |
| 3D3D | (4) | X-RAY CRYSTAL STRUCTURE OF THE PASSENGER DOMAIN OF PLASMID ENCODED TOXIN, AN AUTROTANSPORTER ENTEROTOXIN FROM ENTEROAGGREGATIVE ESCHERICHIA COLI (EAEC) |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF METALLOPROTEINASE-II FROM BACTEROIDES FRAGILIS |
|---|
 | titl: structural and functional diversity of metalloproteinases encoded by the bacteroides fragilis pathogenicity island. |
| 3D3D | (1) | STRUCTURE OF S. POMBE PCT1 RNA TRIPHOSPHATASE |
|---|
 | titl: fission yeast rna triphosphatase reads an spt5 ctd code. |
| 3D3D | (1) | STRUCTURE OF S. POMBE PCT1 RNA TRIPHOSPHATASE IN COMPLEX WITH THE SPT5 CTD |
|---|
 | titl: fission yeast rna triphosphatase reads an spt5 ctd code. |
| 3D3D | (2) | STRUCTURE OF THE CYTOMEGALOVIRUS-ENCODED M04 GLYCOPROTEIN |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF ZINC-BOUND SUCCINYL-DIAMINOPIMELATE DESUCCINYLASE FROM NEISSERIA MENINGITIDIS MC58 |
|---|
 | titl: inhibition of the dape-encoded n-succinyl-l,l-diaminopimeli acid desuccinylase from neisseria meningitidis by l-captopril. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF SUCCINYL-DIAMINOPIMELATE DESUCCINYLASE FROM NEISSERIA MENINGITIDIS MC58 IN COMPLEX WITH THE INHIBITOR CAPTOPRIL |
|---|
 | titl: inhibition of the dape-encoded n-succinyl-l,l-diaminopimeli acid desuccinylase from neisseria meningitidis by l-captopril. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF ANTI-23F STREP FAB C05 |
|---|
 | titl: immunoglobulin v-genes evolved to encode amino acids that can contact chemically distinct antigens to facilitate the generation of protective antibodies. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF ANTI-23F STREP FAB C05 WITH RHAMNOSE |
|---|
 | titl: immunoglobulin v-genes evolved to encode amino acids that can contact chemically distinct antigens to facilitate the generation of protective antibodies. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF FAB 3.1 |
|---|
 | titl: alternative recognition of the conserved stem epitope in influenza a virus hemagglutinin by a vh3-30-encoded heterosubtypic antibody. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF FAB 3.1 IN COMPLEX WITH THE 1918 INFLUENZA VIRUS HEMAGGLUTININ |
|---|
 | titl: alternative recognition of the conserved stem epitope in influenza a virus hemagglutinin by a vh3-30-encoded heterosubtypic antibody. |
| 3D3D | (1) | PCE1 GUANYLYLTRANSFERASE BOUND TO SER2/SER5 PHOSPHORYLATED RNA POL II CTD |
|---|
 | titl: how an mrna capping enzyme reads distinct rna polymerase ii and spt5 ctd phosphorylation codes. |
| 3D3D | (1) | PCE1 GUANYLYLTRANSFERASE |
|---|
 | titl: how an mrna capping enzyme reads distinct rna polymerase ii and spt5 ctd phosphorylation codes. |
| 3D3D | (1) | PCE1 GUANYLYLTRANSFERASE BOUND TO SPT5 CTD |
|---|
 | titl: how an mrna capping enzyme reads distinct rna polymerase ii and spt5 ctd phosphorylation codes. |
| 3D3D | (3) | STRUCTURE OF DIMERIC HUMAN GLYCODELIN |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF RV3772 ENCODED AMINOTRANSFERASE |
|---|
 | crystal structure of rv3772 encoded aminotransferase |
| 3D3D | (2) | CRYSTAL STRUCTURE OF RV1600 ENCODED AMINOTRANSFERASE IN COMPLEX WITH PLP-MES FROM MYCOBACTERIUM TUBERCULOSIS |
|---|
| 3D3D | (2) | CRYSTAL STRUCTURE OF RV1600 ENCODED AMINOTRANSFERASE FROM MYCOBACTERIUM TUBERCULOSIS |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF A PENTAFLUORO-PHE INCORPORATED PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C (H258X)FROM STAPHYLOCOCCUS AUREUS |
|---|
 | cation-pi, tim barrel, encoded unnatural amino acid phospholipase, lyase |
| 3D3D | (1) | CRYSTAL STRUCTURE OF GYRASE BOUND TO ITS INHIBITOR YACG |
|---|
 | titl: direct control of type iia topoisomerase activity by a chromosomally encoded regulatory protein. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE C-TERMINAL DOMAIN OF IFRS BOUND WITH 3-IODO-L-PHE AND ATP |
|---|
 | amino acyl-trna synthetases, archaeal proteins, evolution, molecular, genetic code, substrate specificity, ligase |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE C-TERMINAL DOMAIN OF IFRS BOUND WITH 2-(5-BROMOTHIENYL)-L-ALA AND ATP |
|---|
 | amino acyl-trna synthetases, archaeal proteins, evolution, molecular, genetic code, substrate specificity, ligase |
| 3D3D | (1) | CRYSTAL STRUCTURE OF MEGAVIRUS UDP-GLCNAC 4,6-DEHYDRATASE, 5-EPIMERASE MG534 |
|---|
 | titl: giant virus megavirus chilensis encodes the biosynthetic pathway for uncommon acetamido sugars. |
| 3D3D | (1) | N-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH I-BET726 (GSK1324726A) |
|---|
| 3D3D | (1) | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH I-BET726 (GSK1324726A) |
|---|
| 3D3D | (1) | N-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 1-((2R,4S)-2-METHYL-4-(PHENYLAMINO)-6-(4-(PIPERIDIN-1-YLMETHYL)PHENYL)-3,4-DIHYDROQUINOLIN-1(2H)-YL)ETHANONE |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF GAMMA-B CRYSTALLIN EXPRESSED IN E. COLI BASED ON MRNA VARIANT 2 |
|---|
 | titl: two synonymous gene variants encode proteins with identical sequence, but different folding conformations. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF GAMMA-B CRYSTALLIN EXPRESSED IN E. COLI BASED ON MRNA VARIANT 1 |
|---|
 | titl: two synonymous gene variants encode proteins with identical sequence, but different folding conformations. |
| 3D3D | (1) | STRUCTURE OF DEXTRAN GLUCOSIDASE WITH GLUCOSE |
|---|
 | synonym: dextran glucosidase,exo-1,6-alpha-glucosidase, glucodextranase |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A BACTERIAL FUCODIASE IN COMPLEX WITH 1-((1R,2R, 3R,4R,5R,6R)-2,3,4-TRIHYDROXY-5-METHYL-7-AZABICYCLO[4.1.0]HEPTAN-7-YL)ETHAN-1-ONE |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF A BACTERIAL FUCOSIDASE WITH PHENYL((1R,2R,3R,4R, 5R,6R)-2,3,4-TRIHYDROXY-5-METHYL-7-AZABICYCLO[4.1.0]HEPTAN-7-YL) METHANONE |
|---|
| 3D3D | (1) | STRUCTURE OF DEXTRAN GLUCOSIDASE |
|---|
 | synonym: dextran glucosidase,exo-1,6-alpha-glucosidase, glucodextranase |
| 3D3D | (1) | DCTP DEAMINASE-DUTPASE FROM BACILLUS HALODURANS |
|---|
 | titl: bacillus halodurans strain c125 encodes and synthesizes enzymes from both known pathways to form dump directly from cytosine deoxyribonucleotides. |
| 3D3D | (1) | THE CRYSTAL STRUCTURE OF ERVE VIRUS NUCLEOPROTEIN |
|---|
 | titl: structural and functional diversity of nairovirus-encoded nucleoproteins. |
| 3D3D | (1) | THE CRYSTAL STRUCTURE OF ERVE VIRUS NUCLEOPROTEIN |
|---|
 | titl: structural and functional diversity of nairovirus-encoded nucleoproteins. |
| 3D3D | (1) | THE CRYSTAL STRUCTURE OF KUPE VIRUS NUCLEOPROTEIN |
|---|
 | titl: structural and functional diversity of nairovirus-encoded nucleoproteins. |
| 3D3D | (1) | THE CRYSTAL STRUCTURE OF HAZARA VIRUS NUCLEOPROTEIN |
|---|
 | titl: structural and functional diversity of nairovirus-encoded nucleoproteins. |
| 3D3D | (1) | MNEMIOPSIS LEIDYI ML032222A IGLUR LBD GLYCINE COMPLEX |
|---|
 | titl: glycine activated ion channel subunits encoded by ctenophor glutamate receptor genes. |
| 3D3D | (1) | MNEMIOPSIS LEIDYI ML032222A IGLUR LBD COMPLEX WITH ALANINE |
|---|
 | titl: glycine activated ion channel subunits encoded by ctenophor glutamate receptor genes. |
| 3D3D | (1) | MNEMIOPSIS LEIDYI ML032222A IGLUR LBD D-SERINE COMPLEX |
|---|
 | titl: glycine activated ion channel subunits encoded by ctenophor glutamate receptor genes. |
| 3D3D | (1) | PI3K ALPHA LIPID KINASE WITH ACTIVE SITE INHIBITOR |
|---|
 | titl: discovery of a potent class of pi3k alpha inhibitors with unique binding mode via encoded library technology (elt). |
| 3D3D | (1) | MNEMIOPSIS LEIDYI ML032222A IGLUR LBD SERINE COMPLEX |
|---|
 | titl: glycine activated ion channel subunits encoded by ctenophor glutamate receptor genes. |
| 3D3D | (1) | PLEUROBRACHIA BACHEI IGLUR3 LBD GLYCINE COMPLEX |
|---|
 | titl: glycine activated ion channel subunits encoded by ctenophor glutamate receptor genes. |
| 3D3D | (2) | CRYSTAL STRUCTURE OF A NUCLEAR RECEPTOR BINDING FACTOR 2 MIT DOMAIN (NRBF2) FROM HOMO SAPIENS AT 1.50 A RESOLUTION |
|---|
| 3D3D | (1) | PHOTOTOXIC FLUORESCENT PROTEIN KILLERORANGE |
|---|
 | fluorescent protein, genetically encoded photosensitizer, chromophore-assisted light inactivation |
| 3D3D | (2) | CRYSTAL STRUCTURE OF A DJ-1 (PARK7) FROM HOMO SAPIENS AT 1.23 A RESOLUTION |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE C-TERMINAL DOMAIN OF PYLRS MUTANT BOUND WITH 3-BENZOTHIENYL-L-ALANINE AND ATP |
|---|
 | aminoacyl-trna synthetases, archaeal proteins, evolution, molecular, genetic code, substrate specificity, ligase |
| 3D3D | (2) | GENETICALLY ENCODED PHENYL AZIDE PHOTOCHEMISTRY DRIVE POSITIVE AND NEGATIVE FUNCTIONAL MODULATION OF A RED FLUORESCENT PROTEIN |
|---|
| 3D3D | (1) | IRRADIATED STATE OF MCHERRY143AZF |
|---|
 | titl: genetically encoded phenyl azide photochemistry drives positive and negative functional modulation of a red fluorescent protein |
| 3D3D | (1) | CRYSTAL STRUCTURE OF P-ACRYLAMIDO-PHENYLALANINE MODIFIED TEM1 BETA-LACTAMASE FROM ESCHERICHIA COLI : V216ACRF MUTANT |
|---|
 | titl: exploring the potential impact of an expanded genetic code on protein function. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF P-ACRYLAMIDO-PHENYLALANINE MODIFIED TEM1 BETA-LACTAMASE FROM ESCHERICHIA COLI :E166N MUTANT |
|---|
 | titl: exploring the potential impact of an expanded genetic code on protein function. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF CEPHALEXIN BOUND ACYL-ENZYME INTERMEDIATE OF VAL216ACRF MUTANT TEM1 BETA-LACTAMASE FROM ESCHERICHIA COLI: E166N AND V216ACRF MUTANT. |
|---|
 | titl: exploring the potential impact of an expanded genetic code on protein function. |
| 3D3D | (1) | VIRAL CHEMOKINE BINDING PROTEIN R17 ENCODED BY RODENT GAMMAHERPESVIRUS PERU ( RHVP) |
|---|
 | viral chemokine binding protein r17 encoded by rodent gammaherpesvirus peru ( rhvp) |
| 3D3D | (1) | THE CRYSTAL STRUCTURE OF I-SCEI IN COMPLEX WITH ITS TARGET DNA IN THE PRESENCE OF MN |
|---|
 | molecule: intron-encoded endonuclease i-scei |
| 3D3D | (1) | GH20C, BETA-HEXOSAMINIDASE FROM STREPTOCOCCUS PNEUMONIAE IN COMPLEX WITH GAL-PUGNAC |
|---|
 | titl: a second beta-hexosaminidase encoded in the streptococcus pneumoniae genome provides an expanded biochemical ability to degrade host glycans. |
| 3D3D | (1) | GH20C, BETA-HEXOSAMINIDASE FROM STREPTOCOCCUS PNEUMONIAE IN COMPLEX WITH NGT |
|---|
 | titl: a second beta-hexosaminidase encoded in the streptococcus pneumoniae genome provides an expanded biochemical ability to degrade host glycans. |
| 3D3D | (1) | GH20C, BETA-HEXOSAMINIDASE FROM STREPTOCOCCUS PNEUMONIAE IN COMPLEX WITH PUGNAC |
|---|
 | titl: a second beta-hexosaminidase encoded in the streptococcus pneumoniae genome provides an expanded biochemical ability to degrade host glycans. |
| 3D3D | (1) | GH20C, BETA-HEXOSAMINIDASE FROM STREPTOCOCCUS PNEUMONIAE |
|---|
 | titl: a second beta-hexosaminidase encoded in the streptococcus pneumoniae genome provides an expanded biochemical ability to degrade host glycans. |
| 3D3D | (1) | GH20C, BETA-HEXOSAMINIDASE FROM STREPTOCOCCUS PNEUMONIAE IN COMPLEX WITH GAL-NGT |
|---|
 | titl: a second beta-hexosaminidase encoded in the streptococcus pneumoniae genome provides an expanded biochemical ability to degrade host glycans |
| 3D3D | (1) | GH20C, BETA-HEXOSAMINIDASE FROM STREPTOCOCCUS PNEUMONIAE IN COMPLEX WITH GALNAC |
|---|
 | titl: a second beta-hexosaminidase encoded in the streptococcus pneumoniae genome provides an expanded biochemical ability to degrade host glycans. |
| 3D3D | (1) | GH20C, BETA-HEXOSAMINIDASE FROM STREPTOCOCCUS PNEUMONIAE IN COMPLEX WITH GLCNAC |
|---|
 | titl: a second beta-hexosaminidase encoded in the streptococcus pneumoniae genome provides an expanded biochemical ability to degrade host glycans. |
| 3D3D | (1) | THE CRYSTAL STRUCTURE OF ANTI-H4K20ME1_SCFV, 15F11 |
|---|
 | titl: a genetically encoded probe for live-cell imaging of h4k20 monomethylation |
| 3D3D | (1) | SWITCHING GFP FLUORESCENCE USING GENETICALLY ENCODED PHENYL AZIDE CHEMISTRY THROUGH TWO DIFFERENT NON-NATIVE POST-TRANSLATIONAL MODIFICATIONS ROUTES AT THE SAME POSITION. |
|---|
 | switching gfp fluorescence using genetically encoded phenyl azide chemistry through two different non-native post-translational modifications routes at the same position. |
| 3D3D | (1) | SWITCHING GFP FLUORESCENCE USING GENETICALLY ENCODED PHENYL AZIDE CHEMISTRY THROUGH TWO DIFFERENT NON-NATIVE POST-TRANSLATIONAL MODIFICATIONS ROUTES AT THE SAME POSITION. |
|---|
 | switching gfp fluorescence using genetically encoded phenyl azide chemistry through two different non-native post-translational modifications routes at the same position. |
| 3D3D | (1) | LYSINE 120-ACETYLATED P53 DNA BINDING DOMAIN IN A COMPLEX WITH DNA. |
|---|
 | acetylation, transcription factor, post-translational modification, tumor suppressor, dna binding specificity, apoptosis, induced fit, genetic code expansion, transcription |
| 3D3D | (1) | X-RAY CRYSTAL STRUCTURE AT 2.20A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A PYRAZOLOPYRIMIDINONE FRAGMENT AND AN INTERNAL ALDIMINE LINKED PLP. |
|---|
| 3D3D | (1) | X-RAY CRYSTAL STRUCTURE AT 2.20A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A PYRAZOLOPYRIMIDINONE FRAGMENT AND AN INTERNAL ALDIMINE LINKED PLP. |
|---|
| 3D3D | (1) | X-RAY CRYSTAL STRUCTURE AT 2.17A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A TRIAZOLOPYRIMIDINONE COMPOUND AND AN INTERNAL ALDIMINE LINKED PLP COFACTOR. |
|---|
| 3D3D | (1) | X-RAY CRYSTAL STRUCTURE AT 1.86A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A PYRAZOLOPYRIMIDINONE COMPOUND AND AN INTERNAL ALDIMINE LINKED PLP COFACTOR. |
|---|
| 3D3D | (1) | X-RAY CRYSTAL STRUCTURE AT 1.82A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A PYRROLIDINE AMIDE COMPOUND AND AN INTERNAL ALDIMINE LINKED PLP COFACTOR. |
|---|
| 3D3D | (1) | X-RAY CRYSTAL STRUCTURE AT 1.70A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A 4-CHLORO-2-FLUORO SUBSTITUTED PYRAZOLOPYRIMIDINONE COMPOUND AND AN INTERNAL ALDIMINE LINKED PLP COFACTOR. |
|---|
| 3D3D | (2) | CRYSTAL STRUCTURE OF KORA, A PLASMID-ENCODED, GLOBAL TRANSCRIPTION REGULATOR |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF KORA-OPERATOR DNA COMPLEX (KORA-OA) |
|---|
 | titl: flexibility of kora, a plasmid-encoded, global transcriptio regulator, in the presence and the absence of its operator. |
| 3D3D | (2) | CRYSTAL STRUCTURE OF KORA, A PLASMID-ENCODED, GLOBAL TRANSCRIPTION REGULATOR |
|---|
| 3D3D | (1) | MONOCLINIC COMPLEX STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT WITH A BENZOTRIAZOLE-BASED INHIBITOR GENERATED BY CLICK-CHEMISTRY |
|---|
 | auth: r.swider,m.maslyk,j.m.zapico,c.coderch,r.panchuk, n.skorokhyd,a.schnitzler,k.niefind,b.de pascual-teresa, a.ramos |
| 3D3D | (1) | TETRAGONAL COMPLEX STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT WITH A BENZOTRIAZOLE-BASED INHIBITOR GENERATED BY CLICK-CHEMISTRY |
|---|
 | auth: r.swider,m.maslyk,j.m.zapico,c.coderch,r.panchuk, n.skorokhyd,a.schnitzler,k.niefind,b.de pascual-teresa, a.ramos |
| 3D3D | (1) | X-RAY CRYSTAL STRUCTURE AT 1.61A RESOLUTION OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A BIPHENYL PYRROLIDINE ETHER COMPOUND AND AN INTERNAL ALDIMINE LINKED PLP COFACTOR. |
|---|
 | titl: discovery, sar, and x-ray binding mode study of bcatm inhibitors from a novel dna-encoded library. |
| 3D3D | (2) | ISDB NEAT2 BOUND BY CLONE D2-06 |
|---|
 | isdb neat2, igvh1-69, germline encoded, immune system | |
| titl: germline-encoded neutralization of a staphylococcus aureus virulence factor by the human antibody repertoire. |
| 3D3D | (2) | ISDB NEAT2 BOUND BY D4-30 |
|---|
 | isdb, neat2, germline encoded, immune system | |
| titl: germline-encoded neutralization of a staphylococcus aureus virulence factor by the human antibody repertoire. |
| 3D3D | (2) | ISDB NEAT1 BOUND BY CLONE D4-10 |
|---|
 | titl: germline-encoded neutralization of a staphylococcus aureus virulence factor by the human antibody repertoire. | |
| isdb, neat1, germline encoded, immune system |
| 3D3D | (2) | CRYSTAL STRUCTURE OF AN EF-HAND CALCIUM BINDING DOMAIN OF CAP-BINDING PROTEIN COMPLEX-INTERACTING PROTEIN 1 (EFCAB6) FROM HOMO SAPIENS AT 2.00 A RESOLUTION |
|---|
| 3D3D | (1) | SCCMEC TYPE IV CCH - ACTIVE HELICASE |
|---|
 | titl: staphylococcal sccmec elements encode an active mcm-like helicase and thus may be replicative. |
| 3D3D | (1) | HU DNA-BINDING PROTEIN FROM THERMUS THERMOPHILUS |
|---|
 | other_details: dna-binding protein hu from thermus thermophilus. uniprotkb code: dbh_thet8 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF MOUSE TACO1 |
|---|
 | synonym: coiled-coil domain-containing protein 44,translational activator of mitochondrially-encoded cytochrome c oxidase 1 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE SNX27 PDZ DOMAIN BOUND TO THE C-TERMINAL DGKZETA PDZ BINDING MOTIF |
|---|
 | titl: a molecular code for endosomal recycling of phosphorylated cargos by the snx27-retromer complex. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE SNX27 PDZ DOMAIN BOUND TO THE PHOSPHORYLATED C-TERMINAL 5HT4(A)R PDZ BINDING MOTIF |
|---|
 | titl: a molecular code for endosomal recycling of phosphorylated cargos by the snx27-retromer complex. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE SNX27 PDZ DOMAIN BOUND TO THE PHOSPHORYLATED C-TERMINAL LRRC3B PDZ BINDING MOTIF |
|---|
 | titl: a molecular code for endosomal recycling of phosphorylated cargos by the snx27-retromer complex. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE SNX27 PDZ DOMAIN BOUND TO THE C-TERMINAL PHOSPHORYLATED PTHR PDZ BINDING MOTIF |
|---|
 | titl: a molecular code for endosomal recycling of phosphorylated cargos by the snx27-retromer complex. |
| 3D3D | (1) | STRUCTURE OF SDA1 NUCLEASE APOPROTEIN AS AN EGFP FIXED-ARM FUSION |
|---|
 | synonym: phage-encoded extracellular streptodornase d,phage-encoded streptodornase,streptodornase |
| 3D3D | (1) | STRUCTURE OF SDA1 NUCLEASE WITH BOUND ZINC ION |
|---|
 | synonym: phage-encoded extracellular streptodornase d,phage-encoded streptodornase,streptodornase |
| 3D3D | (1) | RNA DEPENDENT RNA POLYMERASE QDE-1 FROM THIELAVIA TERRESTRIS |
|---|
 | titl: functional evolution in orthologous cell-encoded rna-dependent rna polymerases |
| 3D3D | (1) | SIRT2 IN COMPLEX WITH A 13-MER TRIFLUOROACETYLATED RAN PEPTIDE |
|---|
 | hydrolase, sirtuin, kdac, lysine-deacetylase, lysine-acetylation, genetic-code expansion |
| 3D3D | (1) | BETA-GLUCURONIDASE WITH AN ACTIVITY-BASED PROBE (N-ALKYL CYCLOPHELLITOL AZIRIDINE) BOUND |
|---|
 | auth: l.wu,j.jiang,y.jin,w.w.kallemeijn,c.l.kuo,m.artola,w.dai, c.van elk,m.van eijk,g.a.van der marel,j.d.c.codee, b.i.florea,j.m.f.g.aerts,h.s.overkleeft,g.j.davies |
| 3D3D | (3) | INHA IN COMPLEX WITH A DNA ENCODED LIBRARY HIT |
|---|
| 3D3D | (3) | INHA IN COMPLEX WITH A DNA ENCODED LIBRARY HIT |
|---|
| 3D3D | (3) | INHA IN COMPLEX WITH A DNA ENCODED LIBRARY HIT |
|---|
| 3D3D | (3) | INHA IN COMPLEX WITH A DNA ENCODED LIBRARY HIT |
|---|
| 3D3D | (3) | INHA IN COMPLEX WITH A DNA ENCODED LIBRARY HIT |
|---|
| 3D3D | (2) | STRUCTURE A OF GROUP II INTRON COMPLEXED WITH ITS REVERSE TRANSCRIPTASE |
|---|
 | molecule: group ii intron-encoded protein ltra | |
| transferase, group ii introns, ribonucleoprotein, intron-encoded protein, retrotransposons and spliceosom |
| 3D3D | (1) | STRUCTURE A OF GROUP II INTRON COMPLEXED WITH ITS REVERSE TRANSCRIPTASE |
|---|
 | transferase, group ii introns, ribonucleoprotein, intron-encoded protein, retrotransposons and spliceosom |
| 3D3D | (1) | STRUCTURE OF T-TYPE PHYCOBILIPROTEIN LYASE CPET FROM PROCHLOROCOCCUS PHAGE P-HM1 |
|---|
 | titl: distinct features of cyanophage-encoded t-type phycobiliprotein lyase phi cpet: the role of auxiliary metabolic genes. |
| 3D3D | (1) | X-RAY CRYSTAL STRUCTURE OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A 2-ARYL BENZIMIDAZOLE COMPOUND AND AN INTERNAL ALDIMINE LINKED PLP COFACTOR |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF RIP1 KINASE WITH A BENZO[B][1,4]OXAZEPIN-4-ONE |
|---|
 | titl: dna-encoded library screening identifies benzo[b][1,4]oxazepin-4-ones as highly potent and monoselective receptor interacting protein 1 kinase inhibitors. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF AGD31B, ALPHA-TRANSGLUCOSYLASE IN GLYCOSIDE HYDROLASE FAMILY 31, IN COMPLEX WITH CYCLOPHELLITOL AZIRIDINE PROBE CF022 |
|---|
 | auth: j.jiang,c.l.kuo,l.wu,c.franke,w.w.kallemeijn,b.i.florea, e.van meel,g.a.van der marel,j.d.codee,r.g.boot,g.j.davies, h.s.overkleeft,j.m.aerts |
| 3D3D | (1) | CRYSTAL STRUCTURE OF AGD31B, ALPHA-TRANSGLUCOSYLASE IN GLYCOSIDE HYDROLASE FAMILY 31, IN COMPLEX WITH CYCLOPHELLITOL AZIRIDINE PROBE CF021 |
|---|
 | auth: j.jiang,c.l.kuo,l.wu,c.franke,w.w.kallemeijn,b.i.florea, e.van meel,g.a.van der marel,j.d.codee,r.g.boot,g.j.davies, h.s.overkleeft,j.m.aerts |
| 3D3D | (1) | CRYSTAL STRUCTURE OF C-TERMINAL VARIANT 1 OF CHAETOMIUM THERMOPHILUM ACETYL-COA CARBOXYLASE |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF C-TERMINAL VARIANT 2 OF CHAETOMIUM THERMOPHILUM ACETYL-COA CARBOXYLASE |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF CD-CT DOMAINS OF CHAETOMIUM THERMOPHILUM ACETYL-COA CARBOXYLASE |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF A DBCCP-VARIANT OF CHAETOMIUM THERMOPHILUM ACETYL-COA CARBOXYLASE |
|---|
| 3D3D | (1) | STRUCTURE OF ORF49 FROM KSHV |
|---|
 | titl: structure of the open reading frame 49 protein encoded by kaposi's sarcoma-associated herpesvirus. |
| 3D3D | (1) | STRUCTURE OF THE SPECTRIN REPEATS 5 AND 6 OF THE PLAKIN DOMAIN OF PLECTIN |
|---|
 | other_details: the residues 819-888 that code for an sh3 domain have been replaced by the gly-ser sequence gsgsg |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A 53BP1 TUDOR DOMAIN IN COMPLEX WITH A UBIQUITIN VARIANT |
|---|
 | titl: a genetically encoded inhibitor of 53bp1 to stimulate homology-based gene editing |
| 3D3D | (1) | CRYO-EM STRUCTURE OF A FULL ARCHAEAL RIBOSOMAL TRANSLATION INITIATION COMPLEX IN THE P-REMOTE CONFORMATION |
|---|
| 3D3D | (1) | CRYO-EM STRUCTURE OF A FULL ARCHAEAL RIBOSOMAL TRANSLATION INITIATION COMPLEX IN THE P-IN CONFORMATION |
|---|
| 3D3D | (1) | CATALYTIC DOMAIN OF POLYSPECIFIC PYRROLYSYL-TRNA SYNTHETASE MUTANT N346A/C348A IN COMPLEX WITH AMPPNP |
|---|
 | titl: genetically encoded fluorophenylalanines enable insights into the recognition of lysine trimethylation by an epigenetic reader. |
| 3D3D | (1) | CATALYTIC DOMAIN OF POLYSPECIFIC PYRROLYSYL-TRNA SYNTHETASE MUTANT Y306A/N346A/C348A/Y384F IN COMPLEX WITH AMPPNP |
|---|
 | titl: genetically encoded fluorophenylalanines enable insights into the recognition of lysine trimethylation by an epigenetic reader. |
| 3D3D | (1) | A GLYCOSIDE HYDROLASE MUTANT WITH AN UNREACTED ACTIVITY BASED PROBE BOUND |
|---|
 | auth: l.wu,j.jiang,y.jin,w.w.kallemeijn,c.l.kuo,m.artola,w.dai, c.van elk,m.van eijk,g.a.van der marel,j.d.c.codee, b.i.florea,j.m.f.g.aerts,h.s.overkleeft,g.j.davies |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HUMAN HEPARANASE, IN COMPLEX WITH GLUCURONIC ACID CONFIGURED AZIRIDINE PROBE JJB355 |
|---|
 | auth: l.wu,j.jiang,y.jin,w.w.kallemeijn,c.l.kuo,m.artola,w.dai, c.van elk,m.van eijk,g.a.van der marel,j.d.c.codee, b.i.florea,j.m.f.g.aerts,h.s.overkleeft,g.j.davies |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HUMAN HEPARANASE NUCLEOPHILE MUTANT (E343Q), IN COMPLEX WITH UNREACTED GLUCURONIC ACID CONFIGURED AZIRIDINE PROBE JJB355 |
|---|
 | auth: l.wu,j.jiang,y.jin,w.w.kallemeijn,c.l.kuo,m.artola,w.dai, c.van elk,m.van eijk,g.a.van der marel,j.d.c.codee, b.i.florea,j.m.f.g.aerts,h.s.overkleeft,g.j.davies |
| 3D3D | (1) | CRYSTAL STRUCTURE OF APO HUMAN PROHEPARANASE |
|---|
 | auth: l.wu,j.jiang,y.jin,w.w.kallemeijn,c.l.kuo,m.artola,w.dai, c.van elk,m.van eijk,g.a.van der marel,j.d.c.codee, b.i.florea,j.m.f.g.aerts,h.s.overkleeft,g.j.davies |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HUMAN PROHEPARANASE, IN COMPLEX WITH GLUCURONIC ACID CONFIGURED AZIRIDINE PROBE JJB355 |
|---|
 | auth: l.wu,j.jiang,y.jin,w.w.kallemeijn,c.l.kuo,m.artola,w.dai, c.van elk,m.van eijk,g.a.van der marel,j.d.c.codee, b.i.florea,j.m.f.g.aerts,h.s.overkleeft,g.j.davies |
| 3D3D | (1) | CRYSTAL STRUCTURE OF P38 ALPHA MAPK14 IN COMPLEX WITH VPC00628 |
|---|
 | titl: novel p38alpha map kinase inhibitors identified from yoctoreactor dna-encoded small molecule library |
| 3D3D | (1) | IN-GEL ACTIVITY-BASED PROTEIN PROFILING OF A CLICKABLE COVALENT ERK 1/2 INHIBITOR |
|---|
 | other_details: cys 171 modified to cme by reaction. one letter code should is the same for both |
| 3D3D | (1) | LYSINE 120-ACETYLATED P53 DNA BINDING DOMAIN IN A COMPLEX WITH THE BAX RESPONSE ELEMENT. |
|---|
 | transcription, acetylation, transcription factor, post-translational modification, tumor suppressor, dna binding specificity, apoptosis, induced fit, genetic code expansion |
| 3D3D | (1) | STRUCTURE OF BACTERIAL 30S-IF1-IF3-MRNA TRANSLATION PRE-INITIATION COMPLEX (STATE-1A) |
|---|
| 3D3D | (1) | THE STRUCTURE OF THE METALLO-BETA-LACTAMASE VIM-2 IN COMPLEX WITH A TRIAZOLYLTHIOACETAMIDE INHIBITOR |
|---|
 | antibiotic resistance, carbapenemases, verona integron-encoded metallo-beta-lactamase 2, vim-2, hydrolase |
| 3D3D | (4) | TUBULIN-DISCODERMOLIDE COMPLEX |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF ALPHA-1,2-MANNOSIDASE FROM CAULOBACTER K31 STRAIN IN COMPLEX WITH 1-DEOXYMANNOJIRIMYCIN |
|---|
 | auth: e.r.van rijssel,a.p.a.janssen,a.males,g.j.davies, g.a.van der marel,h.s.overkleeft,j.d.c.codee |
| 3D3D | (1) | VIM-2_2B. METALLO-BETA-LACTAMASE INHIBITORS BY BIOISOSTERIC REPLACEMENT: PREPARATION, ACTIVITY AND BINDING |
|---|
 | synonym: verona integron-encoded metallo-beta-lactamase |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE PROTEIN KINASE CK2 CATALYTIC SUBUNIT IN COMPLEX WITH PYRAZOLO-PYRIMIDINE MACROCYCLIC LIGAND |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE BCL6 BTB DOMAIN IN COMPLEX WITH PYRAZOLO-PYRIMIDINE LIGAND |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE BCL6 BTB DOMAIN IN COMPLEX WITH PYRAZOLO-PYRIMIDINE MACROCYCLIC LIGAND |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE BCL6 BTB DOMAIN IN COMPLEX WITH PYRAZOLO-PYRIMIDINE LIGAND |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE BCL6 BTB DOMAIN IN COMPLEX WITH PYRAZOLO-PYRIMIDINE LIGAND |
|---|
| 3D3D | (1) | THERMOTOGA MARITIMA FAMILY 1 GLYCOSIDE HYDROLASE COMPLEXED WITH CARBA-CYCLOPHELLITOL TRANSITION STATE MIMIC |
|---|
| 3D3D | (1) | THERMOTOGA MARITIMA FAMILY 1 GLYCOSIDE HYDROLASE COMPLEXED WITH A CYCLOPHELLITOL ANALOGUE TRANSITION STATE MIMIC |
|---|
| 3D3D | (1) | CRYSTAL STRUCTURE OF STAPHYLOCOCCUS EPIDERMIDIS AAP G511-SPACER-G513 (CONSENSUS G5-SPACER-CONSENSUS G5) |
|---|
 | titl: functional consequences of b-repeat sequence variation in the staphylococcal biofilm protein aap: deciphering the assembly code. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF STAPHYLOCOCCUS EPIDERMIDIS AAP G58-SPACER-G513* (VARIANT G5-SPACER-VARIANT G5) |
|---|
 | titl: functional consequences of b-repeat sequence variation in the staphylococcal biofilm protein aap: deciphering the assembly code. |
| 3D3D | (1) | CRYSTAL STRUCTURE OF STAPHYLOCOCCUS EPIDERMIDIS AAP G58-SPACER-G513 (VARIANT G5-SPACER-CONSENSUS G5) |
|---|
 | titl: functional consequences of b-repeat sequence variation in the staphylococcal biofilm protein aap: deciphering the assembly code. |
| 3D3D | (3) | DISCOVERY OF A POTENT BTK INHIBITOR WITH A NOVEL BINDING MODE USING PARALLEL SELECTIONS WITH A DNA-ENCODED CHEMICAL LIBRARY |
|---|
|
|