
|
|
| 3D3D | (1) | D95A OXIDIZED FLAVODOXIN MUTANT FROM D. VULGARIS |
|---|
 | pmid: 12206666 |
| 3D3D | (1) | D95A HYDROQUINONE FLAVODOXIN MUTANT FROM D. VULGARIS |
|---|
 | pmid: 12206666 |
| 3D3D | (1) | D95A SEMIQUINONE FLAVODOXIN MUTANT FROM D. VULGARIS |
|---|
 | pmid: 12206666 |
| 3D3D | (1) | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT POSITIONED AT A TEMPLATE-PRIMER JUNCTION, NMR, 6 STRUCTURES |
|---|
 | doi: 10.1021/bi972206r |
| 3D3D | (1) | D95E HYDROQUINONE FLAVODOXIN MUTANT FROM D. VULGARIS |
|---|
 | pmid: 12206666 |
| 3D3D | (1) | D95E OXIDIZED FLAVODOXIN MUTANT FROM D. VULGARIS |
|---|
 | pmid: 12206666 |
| 3D3D | (1) | FIRST STRUCTURAL EVIDENCE OF THE INHIBITION OF PHOSPHOLIPASE A2 BY ARISTOLOCHIC ACID: CRYSTAL STRUCTURE OF A COMPLEX FORMED BETWEEN PHOSPHOLIPASE A2 AND ARISTOLOCHIC ACID |
|---|
 | pmid: 12206661 |
| 3D3D | (1) | HYDROGENASE MATURATION PROTEIN HYPF "ACYLPHOSPHATASE-LIKE" N-TERMINAL DOMAIN (HYPF-ACP) IN COMPLEX WITH SULFATE |
|---|
 | pmid: 12206761 |
| 3D3D | (1) | HYDROGENASE MATURATION PROTEIN HYPF "ACYLPHOSPHATASE-LIKE" N-TERMINAL DOMAIN (HYPF-ACP) IN COMPLEX WITH A SUBSTRATE. CRYSTAL GROWN IN THE PRESENCE OF CARBAMOYLPHOSPHATE |
|---|
 | pmid: 12206761 |
| 3D3D | (1) | LACTOSE AND MES-LIGANDED CONGERIN II |
|---|
 | pmid: 12206768 |
| 3D3D | (1) | LACTOSE-LIGANDED CONGERIN II |
|---|
 | pmid: 12206768 |
| 3D3D | (1) | LIGAND FREE CONGERIN II |
|---|
 | pmid: 12206768 |
| 3D3D | (1) | MES-LIGANDED CONGERIN II |
|---|
 | pmid: 12206768 |
| 3D3D | (1) | RUBISCO FROM GALDIERIA PARTITA |
|---|
 | pmid: 12220629 |
| 3D3D | (1) | ENZYME-SUBSTRATE COMPLEX OF PYRIDOXINE 5'-PHOSPHATE SYNTHASE |
|---|
 | pmid: 12206776 |
| 3D3D | (1) | ENZYME-ANALOGUE SUBSTRATE COMPLEX OF PYRIDOXINE 5'-PHOSPHATE SYNTHASE |
|---|
 | pmid: 12206776 |
| 3D3D | (1) | ENZYME-PHOSPHATE COMPLEX OF PYRIDOXINE 5'-PHOSPHATE SYNTHASE |
|---|
 | pmid: 12206776 |
| 3D3D | (1) | ENZYME-PHOSPHATE2 COMPLEX OF PYRIDOXINE 5'-PHOSPHATE SYNTHASE |
|---|
 | pmid: 12206776 |
| 3D3D | (1) | CRYSTAL STRUCTURE ANALYSIS OF A REDOX-SENSITIVE GREEN FLUORESCENT PROTEIN VARIANT IN A REDUCED FORM |
|---|
 | pmid: 14722062 |
| 3D3D | (1) | CRYSTAL STRUCTURE ANALYSIS OF A REDOX-SENSITIVE GREEN FLUORESCENT PROTEIN VARIANT IN A OXIDIZED FORM |
|---|
 | pmid: 14722062 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF 2,3-DIHYDROXYBIPHENYAL DIOXYGENASE (BPHC) AT 1.45 A RESOLUTION |
|---|
 | pmid: 12206778 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF 2,3-DIHYDROXYBIPHENYL DIOXYGENASE (BPHC) IN COMPLEX WITH 2,3-DIHYDROXYBIPHENYL AT 1.45 A RESOLUTION |
|---|
 | pmid: 12206778 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF BPHC-2,3-DIHYDROXYBIPHENYL-NO COMPLEX |
|---|
 | pmid: 12206778 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF 2,3-DIHYDROXYBIPHENYL DIOXYGENASE (BPHC) IN COMPLEX WITH 2,3-DIHYDROXYBIPHENYL AT 2.0A RESOLUTION |
|---|
 | pmid: 12206778 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE HIS145ALA MUTANT OF 2,3-DIHYDROXYBIPHENY DIOXYGENASE (BPHC) |
|---|
 | pmid: 12206778 |
| 3D3D | (1) | THE HIS145ALA MUTANT OF 2,3-DIHYDROXYBIPHENYL DIOXYGENASE IN COMPLEX WITH 2,3-DIHYDROXYBIPHENYL |
|---|
 | pmid: 12206778 |
| 3D3D | (1) | STRUCTURE OF BPTI MUTANT A16V |
|---|
 | pmid: 12206780 |
| 3D3D | (1) | STRUCTURE OF BPTI_8A MUTANT |
|---|
 | pmid: 12206780 |
| 3D3D | (1) | ZEBULARINE: A NOVEL DNA METHYLATION INHIBITOR THAT FORMS A COVALENT COMPLEX WITH DNA METHYLTRANSFERASE |
|---|
 | pmid: 12206775 |
| 3D3D | (1) | BACTERIORHODOPSIN K INTERMEDIATE AT 1.43 A RESOLUTION |
|---|
 | pmid: 12206785 |
| 3D3D | (1) | BACTERIORHODOPSIN/LIPID COMPLEX AT 1.47 A RESOLUTION |
|---|
 | pmid: 12206785 |
| 3D3D | (1) | BACTERIORHODOPSIN M1 INTERMEDIATE AT 1.43 A RESOLUTION |
|---|
 | pmid: 12206786 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF JACALIN-T-ANTIGEN COMPLEX |
|---|
 | pmid: 12206779 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HUMAN INTERLEUKIN-2 |
|---|
 | pmid: 12582206 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HUMAN IL-2 COMPLEXED WITH (R)-N-[2-[1-(AMINOIMINOMETHYL)-3-PIPERIDINYL]-1-OXOETHYL]-4-(PHENYLETHYNYL)-L-PHENYLALANINE METHYL ESTER |
|---|
 | pmid: 12582206 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HUMAN INTERLEUKIN-2 COMPLEXED WITH SP-1985 |
|---|
 | pmid: 12582206 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HUMAN INTERLEUKIN-2 Y31C COVALENTLY MODIFIED AT C31 WITH (1H-INDOL-3-YL)-(2-MERCAPTO-ETHOXYIMINO)-ACETIC ACID |
|---|
 | pmid: 12582206 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HUMAN INTERLEUKIN-2 K43C COVALENTLY MODIFIED AT C43 WITH 2-[2-(2-CYCLOHEXYL-2-GUANIDINO-ACETYLAMINO)-ACETYLAMINO]-N-(3-MERCAPTO-PROPYL)-PROPIONAMIDE |
|---|
 | pmid: 12582206 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HUMAN INTERLEUKIN-2 |
|---|
 | pmid: 12582206 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF BETA-SECRETASE COMPLEXED WITH INHIBITOR OM00-3 |
|---|
 | pmid: 12206667 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF CALCINEURIN-CYCLOPHILIN-CYCLOSPORIN SHOWS COMMON BUT DISTINCT RECOGNITION OF IMMUNOPHILIN-DRUG COMPLEXES |
|---|
 | doi: 10.1073/pnas.192206699 |
| 3D3D | (1) | CRYSTAL STRUCTURES OF THE NUCLEASE DOMAIN OF COLE7/IM7 IN COMPLEX WITH A PHOSPHATE ION AND A ZINC ION |
|---|
 | doi: 10.1110/ps.0220602 |
| 3D3D | (1) | SOLUTION STRUCTURE OF A TREFOIL-MOTIF-CONTAINING CELL GROWTH FACTOR, PORCINE SPASMOLYTIC PROTEIN |
|---|
 | doi: 10.1073/pnas.91.6.2206; ref: proc.natl.acad.sci.usa v. 91 2206 1994 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PYRIDOXAL KINASE COMPLEXED WITH AMP-PCP AND PYRIDOXAMINE |
|---|
 | pmid: 14722069 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PYRIDOXAL KINASE COMPLEXED WITH ADP AND PLP |
|---|
 | pmid: 14722069 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PYRIDOXAL KINASE COMPLEXED WITH ADP |
|---|
 | pmid: 14722069 |
| 3D3D | (1) | REDUCED FERREDOXIN 6 FROM RHODOBACTER CAPSULATUS |
|---|
 | pmid: 16402206 |
| 3D3D | (2) | CRYSTAL STRUCTURE OF AN ACYL-COA N-ACYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA |
|---|
 | midwest center for structural genomics, mcsg, structural genomics, protein structure initiative, psi, apc22065, transferase | |
| synonym: acyl-coa n-acyltransferase, apc22065 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF AMIDOHYDROROLASE II; NORTHEAST STRUCTURAL GENOMICS TARGET LPR24 |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF P-COUMARIC ACID DECARBOXYLASE (NP_786857.1) FROM LACTOBACILLUS PLANTARUM AT 1.70 A RESOLUTION |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | SOLUTION NMR STRUCTURE OF THE UPF0291 PROTEIN YNZC FROM BACILLUS SUBTILIS. NORTHEAST STRUCTURAL GENOMICS TARGET SR384. |
|---|
 | doi: 10.1002/prot.22064 |
| 3D3D | (1) | X-RAY CRYSTAL STRUCTURE OF PROTEIN PFL_3262 FROM PSEUDOMONAS FLUORESCENS. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET PLR14. |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | 3-METHYLADENINE DNA-GLYCOSYLASE FROM ARCHAEOGLOBUS FULGIDUS |
|---|
 | ref: embo j. v. 26 2206 2007 |
| 3D3D | (1) | 3-METHYLADENINE DNA-GLYCOSYLASE FROM ARCHAEOGLOBUS FULGIDUS |
|---|
 | ref: embo j. v. 26 2206 2007 |
| 3D3D | (1) | SOLUTION NMR STRUCTURE OF THE FOLDED N-TERMINAL FRAGMENT OF UPF0291 PROTEIN YNZC FROM BACILLUS SUBTILIS. NORTHEAST STRUCTURAL GENOMICS TARGET SR384-1-46 |
|---|
 | doi: 10.1002/prot.22064 |
| 3D3D | (1) | NMR STRUCTURE OF HFN14 |
|---|
 | doi: 10.1111/febs.12206 |
| 3D3D | (1) | SOLUTION NMR STRUCTURE OF XENOPUS FN14 |
|---|
 | doi: 10.1111/febs.12206 |
| 3D3D | (1) | SOLUTION NMR STRUCTURE OF BCMA |
|---|
 | doi: 10.1111/febs.12206 |
| 3D3D | (1) | SOLUTION STRUCTURE OF THE THAP-ZINC FINGER DOMAIN 1-81 FROM THE CELL GROWTH SUPPRESSOR HUMAN THAP11 PROTEIN |
|---|
 | gene: thap11, hrihfb2206 |
| 3D3D | (1) | SOLUTION STRUCTURE OF THE C DOMAIN OF RV0899(D236A) FROM MYCOBACTERIUM TUBERCULOSIS |
|---|
 | pmid: 22206986 |
| 3D3D | (1) | SOLUTION STRUCTURE OF THE C DOMAIN OF RV0899 FROM MYCOBACTERIUM TUBERCULOSIS |
|---|
 | pmid: 22206986 |
| 3D3D | (1) | STRUCTURAL BASIS OF THE NON-CODING RNA RSMZ ACTING AS PROTEIN SPONGE: CONFORMER L OF RSMZ(1-72)/RSME(DIMER) 1TO3 COMPLEX |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | STRUCTURAL BASIS OF THE NON-CODING RNA RSMZ ACTING AS PROTEIN SPONGE: CONFORMER R OF RSMZ(1-72)/RSME(DIMER) 1TO3 COMPLEX |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | THE SOLUTION STRUCTURE OF SKINT-1, A CRITICAL DETERMINANT OF DENDRITIC EPIDERMAL GAMMA-DELTA T CELL SELECTION |
|---|
 | doi: 10.1074/jbc.m116.722066 |
| 3D3D | (1) | PLTL-HOLO |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | PLTL-PYRROLYL |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | X-RAY STRUCTURE OF THE PUTATIVE SE BINDING PROTEIN FROM PSEUDOMONAS FLUORESCENS. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET PLR6. |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | T. BRUCEI FARNESYL DIPHOSPHATE SYNTHASE COMPLEXED WITH BISPHOSPHONATE BPH-210 |
|---|
 | doi: 10.1002/prot.22066 |
| 3D3D | (1) | A FERREDOXIN-LIKE METALLO-BETA-LACTAMASE SUPERFAMILY PROTEIN FROM THERMOANAEROBACTER TENGCONGENSIS |
|---|
 | doi: 10.1002/prot.22069 |
| 3D3D | (1) | STRUCTURE OF THE CRO PROTEIN FROM PROPHAGE PFL 6 IN PSEUDOMONAS FLUORESCENS PF-5 |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF Q88U62_LACPL FROM LACTOBACILLUS PLANTARUM. NORTHEAST STRUCTURAL GENOMICS TARGET LPR71 |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE Q4KFT4_PSEF5 PROTEIN FROM PSEUDOMONAS FLUORESCENS. NESG TARGET PLR1 |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | NMR SOLUTION STRUCTURE OF NAPD IN COMPLEX WITH NAPA1-35 SIGNAL PEPTIDE |
|---|
 | gene: napa, b2206, jw2194 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A PUTATIVE TENA-LIKE THIAMINASE (TENA-1, SSO2206) FROM SULFOLOBUS SOLFATARICUS P2 AT 1.50 A RESOLUTION |
|---|
 | crystal structure of a putative tena-like thiaminase (tena-1, sso2206) from sulfolobus solfataricus p2 at 1.50 a resolution |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PUTATIVE DINUCLEOTIDE-BINDING OXIDOREDUCTASE (NP_786167.1) FROM LACTOBACILLUS PLANTARUM AT 1.60 A RESOLUTION |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE THYMIDYLATE SYNTHASE K48Q COMPLEXED WITH DUMP |
|---|
 | ref: int.j.biochem.cell biol. v. 40 2206 2008 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE THYMIDYLATE SYNTHASE K48Q COMPLEXED WITH 5NO2DUMP AND BW1843U89 |
|---|
 | ref: int.j.biochem.cell biol. v. 40 2206 2008 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PROBABLE HAD FAMILY HYDROLASE FROM PSEUDOMONAS FLUORESCENS PF-5 WITH BOUND PHOSPHATE |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF GLUTAMATE DEHYDROGENASE FROM PEPTONIPHILUS ASACCHAROLYTICUS |
|---|
 | pmid: 22068154 |
| 3D3D | (1) | S. CEREVISIAE GERANYLGERANYL PYROPHOSPHATE SYNTHASE IN COMPLEX WITH INHIBITOR BPH-210 |
|---|
 | doi: 10.1002/prot.22066 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF LYSN, ALPHA-AMINOADIPATE AMINOTRANSFERASE (COMPLEXED WITH N-(5'-PHOSPHOPYRIDOXYL)-L-GLUTAMATE), FROM THERMUS THERMOPHILUS HB27 |
|---|
 | pmid: 19632206 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE LAMINARINASE CATALYTIC DOMAIN FROM THERMOTOGA MARITIMA MSB8 |
|---|
 | pmid: 22065588 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE LAMINARINASE CATALYTIC DOMAIN FROM THERMOTOGA MARITIMA MSB8 |
|---|
 | pmid: 22065588 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE LAMINARINASE CATALYTIC DOMAIN FROM THERMOTOGA MARITIMA MSB8 IN COMPLEX WITH GLUCONOLACTONE |
|---|
 | pmid: 22065588 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE LAMINARINASE CATALYTIC DOMAIN FROM THERMOTOGA MARITIMA MSB8 IN COMPLEX WITH CETYLTRIMETHYLAMMONIUM BROMIDE |
|---|
 | pmid: 22065588 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE LAMINARINASE CATALYTIC DOMAIN FROM THERMOTOGA MARITIMA MSB8 |
|---|
 | pmid: 22065588 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE THYMIDYLATE SYNTHASE K48Q |
|---|
 | ref: int.j.biochem.cell biol. v. 40 2206 2008 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE PROPHAGE LP1 PROTEIN 11 |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PYRIDOXAMINE 5'-PHOSPHATE OXIDASE-LIKE PROTEIN (NP_783940.1) FROM LACTOBACILLUS PLANTARUM AT 1.55 A RESOLUTION |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A PUTATIVE CARBOXYLESTERASE (LP_1002) FROM LACTOBACILLUS PLANTARUM WCFS1 AT 2.09 A RESOLUTION |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A PUTATIVE CARBOXYLESTERASE (LP_2923) FROM LACTOBACILLUS PLANTARUM WCFS1 AT 1.70 A RESOLUTION |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF MYO-INOSITOL 2-DEHYDROGENASE (NP_786804.1) FROM LACTOBACILLUS PLANTARUM AT 2.40 A RESOLUTION |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A TRANSCRIPTION REGULATOR FROM LACTOBACILLUS PLANTARUM |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A PUTATIVE INOSITOL CATABOLISM PROTEIN IOLE (IOLE, LP_3607) FROM LACTOBACILLUS PLANTARUM WCFS1 AT 1.85 A RESOLUTION |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PUTATIVE TRANSCRIPTION REGULATOR FROM LACTOBACILLUS PLANTARUM |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE LP_1622 PROTEIN FROM LACTOBACILLUS PLANTARUM. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET LPR114 |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF MUCONATE CYCLOISOMERASE FROM PSEUDOMONAS FLUORESCENS |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | GOLGI MANNOSIDASE II D204A CATALYTIC NUCLEOPHILE MUTANT COMPLEX WITH 3ALPHA,6ALPHA-MANNOPENTAOSE |
|---|
 | doi: 10.1073/pnas.0802206105 |
| 3D3D | (1) | GOLGI ALPHA-MANNOSIDASE II (D204A NUCLEOPHILE MUTANT) IN COMPLEX WITH GNMAN5GN |
|---|
 | doi: 10.1073/pnas.0802206105 |
| 3D3D | (1) | GOLGI ALPHA-MANNOSIDASE II (D204A NUCLEOPHILE MUTANT) |
|---|
 | doi: 10.1073/pnas.0802206105 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF LIPASE/ESTERASE (LP_2923) FROM LACTOBACILLUS PLANTARUM. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET LPR108 |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF MUCONATE LACTONIZING ENZYME FROM PSEUDOMONAS FLUORESCENS COMPLEXED WITH MUCONOLACTONE |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE VPS75 HISTONE CHAPERONE |
|---|
 | ref: proc.natl.acad.sci.usa v. 105 12206 2008 |
| 3D3D | (1) | THE CRYSTAL STRUCTURE OF THE PROTEIN WITH UNKNOWN FUNCTION FROM ARCHAEOGLOBUS FULGIDUS |
|---|
 | gene: coad, af_2206 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PUTATIVE N-ACETYLTRANSFERASE FROM LACTOBACILLUS PLANTARUM |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF MUCONATE LACTONIZING ENZYME FROM PSEUDOMONAS FLUORESCENS COMPLEXED WITH MUCONOLACTONE |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF SACCHAROMYCES CEREVISIAE YGR203W, A HOMOLOG OF SINGLE-DOMAIN RHODANESE AND CDC25 PHOSPHATASE CATALYTIC DOMAIN |
|---|
 | pmid: 19382206 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PEPTIDASE M16 INACTIVE DOMAIN FROM PSEUDOMONAS FLUORESCENS. NORTHEAST STRUCTURAL GENOMICS TARGET PLR293L |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A PERIPLASMIC SUGAR-BINDING PROTEIN FROM THE PSEUDOMONAS FLUORESCENS |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE ANALYSIS OF THE 2,3-DIOXYGENASE LAPB FROM PSEUDOMONAS IN THE COMPLEX WITH 4-METHYLCATECHOL |
|---|
 | strain: kctc 22206 |
| 3D3D | (1) | CRYSTAL STRUCTURE ANALYSIS OF THE 2,3-DIOXYGENASE LAPB FROM PSEUDOMONAS IN COMPLEX WITH A PRODUCT |
|---|
 | strain: kctc 22206 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF SIGNAL RECEIVER DOMAIN OF HD DOMAIN-CONTAINING PROTEIN FROM PSEUDOMONAS FLUORESCENS PF-5 |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE AND COMPUTATIONAL ANALYSES PROVIDE INSIGHTS INTO THE CATALYTIC MECHANISM OF 2, 4-DIACETYLPHLOROGLUCINOL HYDROLASE PHLG FROM PSEUDOMONAS FLUORESCENS |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A BENF-LIKE PORIN FROM PSEUDOMONAS FLUORESCENS PF-5 |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PUTATIVE GERANYLTRANSTRANSFERASE FROM PSEUDOMONAS FLUORESCENS PF-5 |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PUTATIVE GERANYLTRANSTRANSFERASE FROM PSEUDOMONAS FLUORESCENS PF-5 COMPLEXED WITH MAGNESIUM |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF GLUTATHIONE S TRANSFERASE FROM PSEUDOMONAS FLUORESCENS |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | STRUCTURE OF STRINGENT STARVATION PROTEIN A HOMOLOG FROM PSEUDOMONAS FLUORESCENS |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF GLUTATHIONE S TRANSFERASE IN COMPLEX WITH GLUTATHIONE FROM PSEUDOMONAS FLUORESCENS |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF GLUTATHIONE S-TRANSFERASE FROM PSEUDOMONAS FLUORESCENS [PF-5] |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PROBABLE HAD FAMILY HYDROLASE FROM PSEUDOMONAS FLUORESCENS PF-5 |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | BINDING OF KINETIN IN THE ACTIVE SITE OF MISTLETOE LECTIN I |
|---|
 | pmid: 22064121 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF POLYPRENYL SYNTHETASE FROM PSEUDOMONAS FLUORESCENS PF-5 COMPLEXED WITH MAGNESIUM AND ISOPRENYL PYROPHOSPHATE |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A EAL DOMAIN OF GGDEF DOMAIN PROTEIN FROM PSEUDOMONAS FLUORESCENS PF |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HUMAN HDAC6 IN COMPLEX WITH UBIQUITIN |
|---|
 | pmid: 22069321 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PROBABLE HAD FAMILY HYDROLASE FROM PSEUDOMONAS FLUORESCENS PF-5 WITH BOUND MG |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | EPITOPE BACKBONE GRAFTING BY COMPUTATIONAL DESIGN FOR IMPROVED PRESENTATION OF LINEAR EPITOPES ON SCAFFOLD PROTEINS |
|---|
 | pmid: 22061265 |
| 3D3D | (1) | EPITOPE BACKBONE GRAFTING BY COMPUTATIONAL DESIGN FOR IMPROVED PRESENTATION OF LINEAR EPITOPES ON SCAFFOLD PROTEINS |
|---|
 | pmid: 22061265 |
| 3D3D | (1) | EPITOPE BACKBONE GRAFTING BY COMPUTATIONAL DESIGN FOR IMPROVED PRESENTATION OF LINEAR EPITOPES ON SCAFFOLD PROTEINS |
|---|
 | pmid: 22061265 |
| 3D3D | (1) | EPITOPE BACKBONE GRAFTING BY COMPUTATIONAL DESIGN FOR IMPROVED PRESENTATION OF LINEAR EPITOPES ON SCAFFOLD PROTEINS |
|---|
 | pmid: 22061265 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE MOUSE MAB 17.2 |
|---|
 | pmid: 22069505 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF MAB 17.2 IN COMPLEX WITH R13 PEPTIDE |
|---|
 | pmid: 22069505 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF S. AUREUS PYRUVATE KINASE |
|---|
 | pmid: 22066782 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF FAAL28 G330W MUTANT FROM MYCOBACTERIUM TUBERCULOSIS |
|---|
 | pmid: 22206988 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF FACL13 FROM MYCOBACTERIUM TUBERCULOSIS |
|---|
 | pmid: 22206988 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF FACL13 FROM MYCOBACTERIUM TUBERCULOSIS IN DIFFERENT SPACE GROUP C2 |
|---|
 | pmid: 22206988 |
| 3D3D | (1) | STRUCTURE OF THE MOUSE CD1D-GLC-DAG-S2-INKT TCR COMPLEX |
|---|
 | pmid: 22069376 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF NURA |
|---|
 | pmid: 22064858 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF NURA WITH MANGANESE |
|---|
 | pmid: 22064858 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF NURA BOUND TO DAMP AND MANGANESE |
|---|
 | pmid: 22064858 |
| 3D3D | (1) | STRUCTURAL AND FUNCTIONAL CHARACTERIZATION OF HELICOBACTER PYLORI DSBG |
|---|
 | pmid: 22062156 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF ANTHRAX PROTECTIVE ANTIGEN (MEMBRANE INSERTION LOOP DELETED) TO 1.45-A RESOLUTION |
|---|
 | pmid: 22063095 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF ANTHRAX PROTECTIVE ANTIGEN (MEMBRANE INSERTION LOOP DELETED) TO 1.7-A RESOLUTION |
|---|
 | pmid: 22063095 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF ANTHRAX PROTECTIVE ANTIGEN (MEMBRANE INSERTION LOOP DELETED) MUTANT S337C N664C TO 2.06-A RESOLUTION |
|---|
 | pmid: 22063095 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF ANTHRAX PROTECTIVE ANTIGEN MUTANT S337C N664C AND DITHIOLACETONE MODIFIED TO 1.8-A RESOLUTION |
|---|
 | pmid: 22063095 |
| 3D3D | (1) | STRUCTURE OF SH3 CHIMERA WITH A TYPE II LIGAND LINKED TO THE CHAIN C-TERMINAL |
|---|
 | pmid: 22066535 |
| 3D3D | (1) | CRYSTAL STRUCTURE AND RNA BINDING PROPERTIES OF THE RRM/ALKB DOMAINS IN HUMAN ABH8, AN ENZYME CATALYZING TRNA HYPERMODIFICATION, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET HR5601B |
|---|
 | pmid: 22065580 |
| 3D3D | (1) | CRYSTAL STRUCTURE AND RNA BINDING PROPERTIES OF THE RRM/ALKB DOMAINS IN HUMAN ABH8, AN ENZYME CATALYZING TRNA HYPERMODIFICATION, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET HR5601B |
|---|
 | pmid: 22065580 |
| 3D3D | (1) | THE CATALYTIC DOMAIN OF HUMAN DEUBIQUITINASE DUBA |
|---|
 | doi: 10.1038/nsmb.2206 |
| 3D3D | (1) | THE CATALYTIC DOMAIN OF HUMAN DEUBIQUITINASE DUBA IN COMPLEX WITH UBIQUITIN ALDEHYDE |
|---|
 | doi: 10.1038/nsmb.2206 |
| 3D3D | (1) | STRUCTURE OF MOUSE VALPHA14VBETA2-MOUSECD1D-ALPHA-GALACTOSYLCERAMIDE |
|---|
 | pmid: 22065767 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A LIGHT CHAIN DOMAIN OF SCALLOP SMOOTH MUSCLE MYOSIN |
|---|
 | pmid: 22067157 |
| 3D3D | (1) | PHOSPHORYLATED LIGHT CHAIN DOMAIN OF SCALLOP SMOOTH MUSCLE MYOSIN |
|---|
 | pmid: 22067157 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HUMAN SEPTIN 7 GTPASE DOMAIN |
|---|
 | pmid: 22064074 |
| 3D3D | (1) | GEF DOMAIN OF DENND 1B IN COMPLEX WITH RAB GTPASE RAB35 |
|---|
 | pmid: 22065758 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HSULT1A1 BOUND TO PAP |
|---|
 | pmid: 22069470 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HSULT1A1 BOUND TO PAP AND 2-NAPHTOL |
|---|
 | pmid: 22069470 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HUMAN SULT1A1 BOUND TO PAP AND 3-CYANO-7-HYDROXYCOUMARIN |
|---|
 | pmid: 22069470 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HUMAN SULT1A1 BOUND TO PAP AND TWO 3-CYANO-7-HYDROXYCOUMARIN |
|---|
 | pmid: 22069470 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF D249G MUTATED HUMAN SULT1A1 BOUND TO PAP AND P-NITROPHENOL |
|---|
 | pmid: 22069470 |
| 3D3D | (1) | 1E6 TCR SPECIFIC FOR HLA-A*0201-ALWGPDPAAA |
|---|
 | doi: 10.1038/ni.2206 |
| 3D3D | (1) | HUMAN HLA-A*0201-ALWGPDPAAA |
|---|
 | doi: 10.1038/ni.2206 |
| 3D3D | (1) | 1E6-A*0201-ALWGPDPAAA COMPLEX, MONOCLINIC |
|---|
 | doi: 10.1038/ni.2206 |
| 3D3D | (1) | 1E6-A*0201-ALWGPDPAAA COMPLEX, TRICLINIC |
|---|
 | doi: 10.1038/ni.2206 |
| 3D3D | (1) | DESIGN, SYNTHESIS AND BIOLOGICAL EVALUATION OF POTENT QUINOLINE AND PYRROLOQUINOLINE AMMOSAMIDE ANALOGUES AS INHIBITORS FOR QUINONE REDUCTASE 2 |
|---|
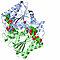 | pmid: 22206487 |
| 3D3D | (1) | DESIGN, SYNTHESIS AND BIOLOGICAL EVALUATION OF POTETENT QUINOLINE AND PYRROLOQUINOLINE AMMOSAMIDE ANALOGUES AS INHIBITORS OF QUINONE REDUCTASE 2 |
|---|
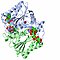 | pmid: 22206487 |
| 3D3D | (1) | CRYSTAL OF PLASMODIUM FALCIPARUM TYROSYL-TRNA SYNTHETASE (PFTYRRS)IN COMPLEX WITH ADENYLATE ANALOG |
|---|
 | pmid: 22068597 |
| 3D3D | (1) | VITAMIN D RECEPTOR COMPLEX WITH A CARBORANE COMPOUND |
|---|
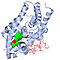 | pmid: 22066785 |
| 3D3D | (1) | VITAMIN D RECEPTOR COMPLEX WITH A CARBORANE COMPOUND |
|---|
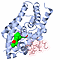 | pmid: 22066785 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF COMPLEXES OF VITAMIN D RECEPTOR LIGAND BINDING DOMAIN WITH LITHOCHOLIC ACID DERIVATIVES |
|---|
 | ref: j.lipid res. v. 54 2206 2013 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF COMPLEXES OF VITAMIN D RECEPTOR LIGAND BINDING DOMAIN WITH LITHOCHOLIC ACID DERIVATIVES |
|---|
 | ref: j.lipid res. v. 54 2206 2013 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF COMPLEXES OF VITAMIN D RECEPTOR LIGAND BINDING DOMAIN WITH LITHOCHOLIC ACID DERIVATIVES |
|---|
 | ref: j.lipid res. v. 54 2206 2013 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF COMPLEXES OF VITAMIN D RECEPTOR LIGAND BINDING DOMAIN WITH LITHOCHOLIC ACID DERIVATIVES |
|---|
 | ref: j.lipid res. v. 54 2206 2013 |
| 3D3D | (1) | STRUCTURE OF THE LECTIN FROM PLATYPODIUM ELEGANS IN COMPLEX WITH A TRIMANNOSIDE |
|---|
 | pmid: 22692206 |
| 3D3D | (1) | STRUCTURE OF THE LECTIN FROM PLATYPODIUM ELEGANS IN COMPLEX WITH HEPTASACCHARIDE |
|---|
 | pmid: 22692206 |
| 3D3D | (1) | PSEUDOMONAS AERUGINOSA PHOSPHORYLCHOLINE PHOSPHATASE. MONOCLINIC FORM |
|---|
 | pmid: 22922065 |
| 3D3D | (1) | PSEUDOMONAS AERUGINOSA PHOSPHORYLCHOLINE PHOSPHATASE. ORTHORHOMBIC FORM |
|---|
 | pmid: 22922065 |
| 3D3D | (1) | COMPLETE CRYSTAL STRUCTURE OF THE CARBOXYLESTERASE CEST-2923 (LP_2923) FROM LACTOBACILLUS PLANTARUM WCFS1 |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | COMPLETE CRYSTAL STRUCTURE OF CARBOXYLESTERASE CEST-2923 FROM LACTOBACILLUS PLANTARUM WCFS1 |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | COMPLETE CRYSTAL STRUCTURE OF CARBOXYLESTERASE CEST-2923 (LP_2923) FROM LACTOBACILLUS PLANTARUM WCFS1 |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | ESTERASE LPEST1 FROM LACTOBACILLUS PLANTARUM WCFS1 |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | ESTERASE LPEST1 FROM LACTOBACILLUS PLANTARUM: NATIVE STRUCTURE |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF CARBOXYLESTERASE LPEST1 FROM LACTOBACILLUS PLANTARUM: HIGH RESOLUTION MODEL |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | THE CRYO-ELECTRON MICROSCOPY STRUCTURE OF THE CORA CHANNEL FROM METHANOCALDOCOCCUS JANNASCHII AT 21.6 ANGSTROM IN LOW MAGNESIUM. |
|---|
 | ref: biochim. biophys. acta v.1848 2206 2015 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE ROP PROTEIN MUTANT D30P/A31G AT RESOLUTION 1.4 RESOLUTION. |
|---|
 | doi: 10.1073/pnas.1322065111 |
| 3D3D | (1) | STRUCTURE OF THE MINIMAL STE5 VWA DOMAIN SUBJECT TO AUTOINHIBITION BY THE STE5 PH DOMAIN |
|---|
 | doi: 10.1126/science.1220683 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF 6-PHOSPHO-BETA-GLUCOSIDASE FROM LACTOBACILLUS PLANTARUM (APO FORM) |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A GLUTATHIONE TRANSFERASE FAMILY MEMBER FROM PSUEDOMONAS FLUORESCENS PF-5, TARGET EFI-900011, WITH BOUND S-(PROPANOIC ACID)-GLUTATHIONE |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF A GLUTATHIONE TRANSFERASE FAMILY MEMBER FROM PSEUDOMONAS FLUORESCENS PF-5, TARGET EFI-900003, WITH TWO GLUTATHIONE BOUND |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | (M)L214G MUTANT OF THE RHODOBACTER SPHAEROIDES REACTION CENTER |
|---|
 | ref: biochemistry v. 52 2206 2013 |
| 3D3D | (1) | (M)L214A MUTANT OF THE RHODOBACTER SPHAEROIDES REACTION CENTER |
|---|
 | ref: biochemistry v. 52 2206 2013 |
| 3D3D | (1) | (M)L214N MUTANT OF THE RHODOBACTER SPHAEROIDES REACTION CENTER |
|---|
 | ref: biochemistry v. 52 2206 2013 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF MITOCHONDRIAL HSP90 (TRAP1) WITH AMPPNP |
|---|
 | pmid: 24462206 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF MITOCHONDRIAL HSP90 (TRAP1) NTD-MIDDLE DOMAIN DIMER WITH AMPPNP |
|---|
 | pmid: 24462206 |
| 3D3D | (1) | STRUCTURE OF MITOCHONDRIAL HSP90 (TRAP1) WITH ADP-ALF4- |
|---|
 | pmid: 24462206 |
| 3D3D | (1) | STRUCTURE OF MITOCHONDRIAL HSP90 (TRAP1) WITH ADP-BEF3 |
|---|
 | pmid: 24462206 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE COMPLEX OF A HYDROXYPROLINE EPIMERASE (TARGET EFI-506499, PSEUDOMONAS FLUORESCENS PF-5) WITH THE INHIBITOR PYRROLE-2-CARBOXYLATE |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE COMPLEX OF A HYDROXYPROLINE EPIMERASE (TARGET EFI-506499, PSEUDOMONAS FLUORESCENS PF-5) WITH TRANS-4-HYDROXY-L-PROLINE |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | SECRETED CHLAMYDIAL PROTEIN PGP3, FULL-LENGTH |
|---|
 | ref: j.biol.chem. v. 288 22068 2013 |
| 3D3D | (1) | SECRETED CHLAMYDIAL PROTEIN PGP3, C-TERMINAL DOMAIN |
|---|
 | ref: j.biol.chem. v. 288 22068 2013 |
| 3D3D | (1) | SECRETED CHLAMYDIAL PROTEIN PGP3, COILED-COIL DELETION |
|---|
 | ref: j.biol.chem. v. 288 22068 2013 |
| 3D3D | (1) | STRUCTURE OF A TYPE VI SECRETION SYSTEM EFFECTOR-IMMUNITY COMPLEX FROM PSEUDOMONAS PROTEGENS |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | THE CRYSTAL STRUCTURE OF AMINO ACID ABC TRANSPORTER SUBSTRATE-BINDING PROTEIN FROM PSEUDOMONAS FLUORESCENS PF-5 |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF BRUTON AGAMMAGLOBULINEMIA TYROSINE KINASE COMPLEXED WITH BMS-809959 AKA 4-TERT-BUTYL-N-[2-ME THYL-3-(6-{[4-(MORPHOLINE-4-CARBONYL)PHENYL]AMINO}-9H- PURIN-2-YL)PHENYL]BENZAMIDE |
|---|
 | ref: bioorg.med.chem.lett. v. 24 2206 2014 |
| 3D3D | (1) | THE CRYSTAL STRUCTURE OF A CLASS V CHITININASE FROM PSEUDOMONAS FLUORESCENS PF-5 |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | THE CRYSTAL STRUCTURE OF ABC TRANSPORTER PERMEASE FROM PSEUDOMONAS FLUORESCENS GROUP |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF LPSPL/LPP2128, LEGIONELLA PNEUMOPHILA SPHINGOSINE-1 PHOSPHATE LYASE |
|---|
 | doi: 10.1073/pnas.1522067113 |
| 3D3D | (1) | UNDA, AN OXYGEN-ACTIVATING, NON-HEME IRON DEPENDENT DESATURASE/DECARBOXYLASE |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | UNDA COMPLEXED WITH 2,3-DODECENOIC ACID |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | UNDA COMPLEXED WITH BETA-HYDROXYDODECANOIC ACID |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | NADH:QUINONE OXIDOREDUCTASE (NDH-II) FROM STAPHYLOCOCCUS AUREUS - HOLOPROTEIN STRUCTURE |
|---|
 | pmid: 26172206 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF CYSTATHIONINE BETA-SYNTHASE FROM LACTOBACILLUS PLANTARUM |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF K42A MUTANT OF CYSTATHIONINE BETA-SYNTHASE FROM LACTOBACILLUS PLANTARUM IN A COMPLEX WITH L-METHIONINE |
|---|
 | organism_taxid: 220668 |
| 3D3D | (1) | STRUCTURE OF THE PENICILLIN-BINDING PROTEIN PONA1 FROM MYCOBACTERIUM TUBERCULOSIS |
|---|
 | ref: febs j. v. 283 2206 2016 |
| 3D3D | (1) | STRUCTURE OF THE PONA1 PROTEIN FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH PENICILLIN V |
|---|
 | ref: febs j. v. 283 2206 2016 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HALOGENASE PLTA |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | STRUCTURE OF MN BOUND DUF89 FROM SACCHAROMYCES CEREVISIAE |
|---|
 | pmid: 27322068 |
| 3D3D | (1) | CYP153D17 FROM SPHINGOMONAS SP. PAMC 26605 |
|---|
 | doi: 10.3390/ijms17122067 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE DEHYDRATASE DOMAIN OF RZXB FROM PSEUDOMONAS FLUORESCENS |
|---|
 | organism_taxid: 220664 |
| 3D3D | (1) | HUMAN GIVD CYTOSOLIC PHOSPHOLIPASE A2 IN COMPLEX WITH METHYL GAMMA-LINOLENYL FLUOROPHOSPHONATE |
|---|
 | pmid: 27220631 |
| 3D3D | (1) | HUMAN GIVD CYTOSOLIC PHOSPHOLIPASE A2 |
|---|
 | pmid: 27220631 |
| 3D3D | (1) | HUMAN GIVD CYTOSOLIC PHOSPHOLIPASE A2 IN COMPLEX WITH METHYL GAMMA-LINOLENYL FLUOROPHOSPHONATE INHIBITOR AND TERBIUM CHLORIDE |
|---|
 | pmid: 27220631 |
| 3D3D | (1) | X-RAY STRUCTURE OF THE COMPLEX BETWEEN BOVINE PANCREATIC RIBONUCLEASE AND PENTACHLOROCARBONYLIRIDATE(III) (4 DAYS OF SOAKING) |
|---|
 | ref: dalton trans v. 45 12206 2016 |
| 3D3D | (1) | X-RAY STRUCTURE OF THE COMPLEX BETWEEN BOVINE PANCREATIC RIBONUCLEASE AND PENTHACHLOROCARBONYLIRIDATE(III) (2 MONTHS OF SOAKING) |
|---|
 | ref: dalton trans v. 45 12206 2016 |
| 3D3D | (1) | STRUCTURE OF THE THERMOSTABILIZED EAAT1 CRYST MUTANT IN COMPLEX WITH L-ASP AND THE ALLOSTERIC INHIBITOR UCPH101 |
|---|
 | doi: 10.1038/nature22064 |
| 3D3D | (1) | STRUCTURE OF THE THERMOSTABILIZED EAAT1 CRYST-II MUTANT IN COMPLEX WITH L-ASP |
|---|
 | doi: 10.1038/nature22064 |
| 3D3D | (1) | STRUCTURE OF THE THERMOSTALILIZED EAAT1 CRYST-II MUTANT IN COMPLEX WITH L-ASP AND THE ALLOSTERIC INHIBITOR UCPH101 |
|---|
 | doi: 10.1038/nature22064 |
| 3D3D | (1) | HELICAL ASSEMBLY OF THE ANBU COMPLEX FROM PSEUDOMONAS AERUGINOSA |
|---|
 | gene: ao946_34255, paerug_e15_london_28_01_14_04292, paerug_p32_london_17_vim_2_10_11_02206 |
| 3D3D | (1) | STRUCTURE OF THE THERMOSTABILIZED EAAT1 CRYST MUTANT IN COMPLEX WITH THE COMPETITITVE INHIBITOR TFB-TBOA AND THE ALLOSTERIC INHIBITOR UCPH101 |
|---|
 | doi: 10.1038/nature22064 |
| 3D3D | (1) | STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS PROTEASOME IN COMPLEX WITH N, C-CAPPED DIPEPTIDE PKS2206 |
|---|
 | structure of mycobacterium tuberculosis proteasome in complex with n, c-capped dipeptide pks2206 |
|
|