|
|
| 3D3D | (1) | RETINAL DEHYDROGENASE TYPE TWO WITH NAD BOUND |
|---|
 | ref: biochemistry v. 38 6003 1999 |
| 3D3D | (1) | PURINE NUCLEOSIDE PHOSPHORYLASE FROM CELLULOMONAS SP. IN COMPLEX WITH 8-IODO-GUANINE |
|---|
 | pmid: 10600382 |
| 3D3D | (1) | RNASE T1 VARIANT WITH ALTERED GUANINE BINDING SEGMENT |
|---|
 | pmid: 10600381 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HUMAN PROFILIN II |
|---|
 | pmid: 10600384 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF CHONDROITINASE B |
|---|
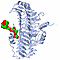 | pmid: 10600383 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF CHONDROITINASE B |
|---|
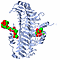 | pmid: 10600383 |
| 3D3D | (1) | 1H NMR SOLUTION STRUCTURE OF CYCLOVIOLACIN O1 |
|---|
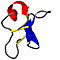 | pmid: 10600388 |
| 3D3D | (1) | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGMO6AATTCGCG) AT 1.6 ANGSTROM |
|---|
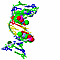 | pmid: 10600380 |
| 3D3D | (1) | PURINE NUCLEOSIDE PHOSPHORYLASE FROM CELLULOMONAS SP. IN COMPLEX WITH PHOSPHATE |
|---|
 | pmid: 10600382 |
| 3D3D | (1) | TRUNCATED HUMAN GROB[5-73], NMR, 20 STRUCTURES |
|---|
 | pmid: 10600366 |
| 3D3D | (1) | METHIONYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI |
|---|
 | pmid: 10600385 |
| 3D3D | (1) | SOLUTION STRUCTURE OF THE HEADPIECE DOMAIN OF CHICKEN VILLIN |
|---|
 | pmid: 10600386 |
| 3D3D | (1) | HISTONE ACETYLTRANSFERASE HPA2 FROM SACCHAROMYCES CEREVISIAE IN COMPLEX WITH ACETYL COENZYME A |
|---|
 | pmid: 10600387 |
| 3D3D | (1) | HISTONE ACETYLTRANSFERASE HPA2 FROM SACCHAROMYCES CEREVISIAE |
|---|
 | pmid: 10600387 |
| 3D3D | (1) | POLIOVIRUS POLYMERASE FULL LENGTH APO STRUCTURE |
|---|
 | doi: 10.1038/sj.emboj.7600357 |
| 3D3D | (1) | POLIOVIRUS POLYMERASE WITH GTP |
|---|
 | doi: 10.1038/sj.emboj.7600357 |
| 3D3D | (1) | POLIOVIRUS POLYMERASE WITH A 68 RESIDUE N-TERMINAL TRUNCATION |
|---|
 | doi: 10.1038/sj.emboj.7600357 |
| 3D3D | (1) | PROPIONIBACTERIUM SHERMANII TRANSCARBOXYLASE 5S SUBUNIT |
|---|
 | doi: 10.1038/sj.emboj.7600373 |
| 3D3D | (1) | PROPIONIBACTERIUM SHERMANII TRANSCARBOXYLASE 5S SUBUNIT BOUND TO OXALOACETATE |
|---|
 | doi: 10.1038/sj.emboj.7600373 |
| 3D3D | (1) | PROPIONIBACTERIUM SHERMANII TRANSCARBOXYLASE 5S SUBUNIT BOUND TO PYRUVIC ACID |
|---|
 | doi: 10.1038/sj.emboj.7600373 |
| 3D3D | (1) | PROPIONIBACTERIUM SHERMANII TRANSCARBOXYLASE 5S SUBUNIT BOUND TO 2-KETOBUTYRIC ACID |
|---|
 | doi: 10.1038/sj.emboj.7600373 |
| 3D3D | (1) | COMPLEX OF LMO4 LIM DOMAINS 1 AND 2 WITH THE LDB1 LID DOMAIN |
|---|
 | doi: 10.1038/sj.emboj.7600376 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE GLYCOGEN SYNTHASE FROM A. TUMEFACIENS IN COMPLEX WITH ADP |
|---|
 | doi: 10.1038/sj.emboj.7600324 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE GLYCOGEN SYNTHASE FROM AGROBACTERIUM TUMEFACIENS (NON-COMPLEXED FORM) |
|---|
 | doi: 10.1038/sj.emboj.7600324 |
| 3D3D | (1) | PROPIONIBACTERIUM SHERMANII TRANSCARBOXYLASE 5S SUBUNIT A59T |
|---|
 | doi: 10.1038/sj.emboj.7600373 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE TNSA/TNSC(504-555) COMPLEX |
|---|
 | doi: 10.1038/sj.emboj.7600311 |
| 3D3D | (1) | FORMYL-COA TRANFERASE MUTANT ASP169 TO SER |
|---|
 | ref: j.biol.chem. v. 279 36003 2004 |
| 3D3D | (1) | STRUCTURAL BASIS OF ACTIN SEQUESTRATION BY THYMOSIN-B4: IMPLICATIONS FOR ARP2/3 ACTIVATION |
|---|
 | doi: 10.1038/sj.emboj.7600372 |
| 3D3D | (1) | FORMYL-COA TRANSFERASE IN COMPLEX WITH OXALYL-COA |
|---|
 | ref: j.biol.chem. v. 279 36003 2004 |
| 3D3D | (1) | T7 DNA POLYMERASE TERNARY COMPLEX WITH DCTP AT THE INSERTION SITE. |
|---|
 | doi: 10.1038/sj.emboj.7600354 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF OUTER MEMBRANE ENZYME PAGP |
|---|
 | doi: 10.1038/sj.emboj.7600320 |
| 3D3D | (1) | T7 DNA POLYMERASE TERNARY COMPLEX WITH 8 OXO GUANOSINE AND DDCTP AT THE INSERTION SITE |
|---|
 | doi: 10.1038/sj.emboj.7600354 |
| 3D3D | (1) | T7 DNA POLYMERASE BINARY COMPLEX WITH 8 OXO GUANOSINE IN THE TEMPLATING STRAND |
|---|
 | doi: 10.1038/sj.emboj.7600354 |
| 3D3D | (1) | T7 DNA POLYMERASE TERNARY COMPLEX WITH 8 OXO GUANOSINE AND DAMP AT THE ELONGATION SITE |
|---|
 | doi: 10.1038/sj.emboj.7600354 |
| 3D3D | (1) | T7 DNA POLYMERASE TERNARY COMPLEX WITH 8 OXO GUANOSINE AND DCMP AT THE ELONGATION SITE |
|---|
 | doi: 10.1038/sj.emboj.7600354 |
| 3D3D | (1) | SOLUTION STRUCTURE OF T4 BACTERIPHAGE ASIA MONOMER |
|---|
 | doi: 10.1038/sj.emboj.7600312 |
| 3D3D | (1) | T4 ASIA BOUND TO SIGMA70 REGION 4 |
|---|
 | doi: 10.1038/sj.emboj.7600312 |
| 3D3D | (1) | TSX STRUCTURE COMPLEXED WITH THYMIDINE |
|---|
 | doi: 10.1038/sj.emboj.7600330 |
| 3D3D | (1) | TSX STRUCTURE |
|---|
 | doi: 10.1038/sj.emboj.7600330 |
| 3D3D | (1) | TSX STRUCTURE COMPLEXED WITH URIDINE |
|---|
 | doi: 10.1038/sj.emboj.7600330 |
| 3D3D | (1) | POLIOVIRUS POLYMERASE G1A MUTANT |
|---|
 | doi: 10.1038/sj.emboj.7600357 |
| 3D3D | (1) | STRUCTURE OF THE BIFUNCTIONAL AND GOLGI ASSOCIATED FORMIMINOTRANSFERASE CYCLODEAMINASE OCTAMER |
|---|
 | doi: 10.1038/sj.emboj.7600327 |
| 3D3D | (1) | TRANSTHYRETIN-V/122/I CARDIOMYOPATHIC MUTANT |
|---|
 | doi: 10.1107/s0907444996003307 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF CSL BOUND TO DNA |
|---|
 | doi: 10.1038/sj.emboj.7600349 |
| 3D3D | (1) | SOLUTION STRUCTURE OF SIGMA A REGION 4 FROM THERMOTOGA MARITIMA |
|---|
 | doi: 10.1038/sj.emboj.7600312 |
| 3D3D | (1) | TRIGLYCINE VARIANT OF THE ARNO PLECKSTRIN HOMOLOGY DOMAIN IN COMPLEX WITH INS(1,3,4,5)P4 |
|---|
 | doi: 10.1038/sj.emboj.7600388 |
| 3D3D | (1) | TRIGLYCINE VARIANT OF THE ARNO PLECKSTRIN HOMOLOGY DOMAIN IN COMPLEX WITH INS(1,4,5)P3 |
|---|
 | doi: 10.1038/sj.emboj.7600388 |
| 3D3D | (1) | TRIGLYCINE VARIANT OF THE GRP1 PLECKSTRIN HOMOLOGY DOMAIN UNLIGANDED |
|---|
 | doi: 10.1038/sj.emboj.7600388 |
| 3D3D | (1) | PROPIONIBACTERIUM SHERMANII TRANSCARBOXYLASE 5S SUBUNIT, MET186ILE |
|---|
 | doi: 10.1038/sj.emboj.7600373 |
| 3D3D | (1) | REGULATION OF CYTOLYSIN EXPRESSION BY ENTEROCOCCUS FAECALIS: ROLE OF CYLR2 |
|---|
 | doi: 10.1038/sj.emboj.7600367 |
| 3D3D | (1) | FORMYL-COA TRANSFERASE MUTANT ASP169 TO ALA |
|---|
 | ref: j.biol.chem. v. 279 36003 2004 |
| 3D3D | (1) | FORMYL-COA TRANSFERASE MUTANT ASP169 TO GLU |
|---|
 | ref: j.biol.chem. v. 279 36003 2004 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE PDK1 PLECKSTRIN HOMOLOGY (PH) DOMAIN BOUND TO INOSITOL (1,3,4,5)-TETRAKISPHOSPHATE |
|---|
 | doi: 10.1038/sj.emboj.7600379 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE PDK1 PLECKSTRIN HOMOLOGY (PH) DOMAIN BOUND TO DIC4-PHOSPHATIDYLINOSITOL (3,4,5)-TRISPHOSPHATE |
|---|
 | doi: 10.1038/sj.emboj.7600379 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE PDK1 PLECKSTRIN HOMOLOGY (PH) DOMAIN |
|---|
 | doi: 10.1038/sj.emboj.7600379 |
| 3D3D | (1) | HUMAN CDK2 IN COMPLEX WITH OLOMOUCINE II, A NOVEL 2,6,9-TRISUBSTITUTED PURINE CYCLIN-DEPENDENT KINASE INHIBITOR |
|---|
 | pmid: 16003486 |
| 3D3D | (1) | CRYSTAL STRUCUTRE OF ECHINOMYCIN-(ACGTACGT)2 SOLVED BY SAD |
|---|
 | doi: 10.1107/s0907444906003763 |
| 3D3D | (1) | STRUCTURE OF HUMAN ARTEMIN |
|---|
 | doi: 10.1021/bi060035x |
| 3D3D | (1) | STRUCTURE OF THE RB C-TERMINAL DOMAIN BOUND TO AN E2F1-DP1 HETERODIMER |
|---|
 | pmid: 16360038 |
| 3D3D | (1) | SPECIFIC BINDING OF NON-STEROIDAL ANTI-INFLAMMATORY DRUGS (NSAIDS) TO PHOSPHOLIPASE A2: CRYSTAL STRUCTURE OF THE COMPLEX FORMED BETWEEN PHOSPHOLIPASE A2 AND DICLOFENAC AT 2.7 A RESOLUTION: |
|---|
 | doi: 10.1107/s0907444906003660 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE HOLO FORM OF GLUTAREDOXIN C1 FROM POPULUS TREMULA X TREMULOIDES |
|---|
 | pmid: 17460036 |
| 3D3D | (1) | X-RAY STRUCTURE OF HUMAN CDK6-VCYCLIN IN COMPLEX WITH THE INHIBITOR PD0332991 |
|---|
 | doi: 10.1021/jm0600388 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF E. COLI. METHIONINE AMINO PEPTIDASE IN COMPLEX WITH 5-(2-(TRIFLUOROMETHYL)PHENYL)FURAN-2-CARBOXYLIC ACID |
|---|
 | doi: 10.1107/s0907444906003878 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF METHIONINE AMINOPEPTIDASE IN COMPLEX WITH 5-(2,5-DICHLOROPHENYL)FURAN-2-CARBOXYLIC ACID |
|---|
 | doi: 10.1107/s0907444906003878 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF METHIONINE AMINO PEPTIDASE IN COMPLEX WITH N-CYCLOPENTYL-N-(THIAZOL-2-YL)OXALAMIDE |
|---|
 | doi: 10.1107/s0907444906003878 |
| 3D3D | (1) | X-RAY STRUCTURE OF HUMAN CDK6-VCYCLINWITH THE INHIBITOR AMINOPURVALANOL |
|---|
 | doi: 10.1021/jm0600388 |
| 3D3D | (1) | SOLUTION STRUCTURE OF THE SPLIT PH DOMAIN IN PHOSPHOLIPASE C-GAMMA1 |
|---|
 | doi: 10.1074/jbc.m600336200 |
| 3D3D | (1) | SOLUTION STRUCTURE OF AN EGF-LIKE DOMAIN FROM THE PLASMODIUM FALCIPARUM MEROZOITE SURFACE PROTEIN 1 |
|---|
 | doi: 10.1002/cbic.200600357 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE HOMO SAPIENS CYTOPLASMIC RIBOSOMAL DECODING SITE COMPLEXED WITH APRAMYCIN |
|---|
 | doi: 10.1002/anie.200600354 |
| 3D3D | (1) | CRYSTALLOGRAPHIC STRUCTURE OF THE MOLYBDATE-BINDING PROTEIN OF XANTHOMONAS CITRI AT 1.7 ANG RESOLUTION BOUND TO MOLYBDATE |
|---|
 | doi: 10.1107/s1744309106003812 |
| 3D3D | (1) | PHAGE-SELECTED HOMEODOMAIN BOUND TO UNMODIFIED DNA |
|---|
 | doi: 10.1021/cb6003756 |
| 3D3D | (1) | PHAGE SELECTED HOMEODOMAIN BOUND TO MODIFIED DNA |
|---|
 | doi: 10.1021/cb6003756 |
| 3D3D | (1) | INTER-SUBUNIT SIGNALING IN GSAM |
|---|
 | doi: 10.1073/pnas.0600306103 |
| 3D3D | (1) | INTER-SUBUNIT SIGNALING IN GSAM |
|---|
 | doi: 10.1073/pnas.0600306103 |
| 3D3D | (1) | INTER-SUBUNIT SIGNALING IN GSAM |
|---|
 | doi: 10.1073/pnas.0600306103 |
| 3D3D | (1) | INTER-SUBUNIT SIGNALING IN GSAM |
|---|
 | doi: 10.1073/pnas.0600306103 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH DHBNH, AN RNASE H INHIBITOR |
|---|
 | doi: 10.1021/cb600303y |
| 3D3D | (1) | CRYSTAL STRUCTURE OF T330S MUTANT OF RV3290C FROM M. TUBERCULOSIS |
|---|
 | pmid: 26003725 |
| 3D3D | (1) | N328A MUTANT OF M. TUBERCULOSIS RV3290C |
|---|
 | pmid: 26003725 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE M. TUBERCULOSIS LYSINE-EPSILON AMINOTRANSFERASE (RV3290C) COMPLEXED TO AN INHIBITOR |
|---|
 | pmid: 26003725 |
| 3D3D | (1) | E243 MUTANT OF M. TUBERCULOSIS RV3290C |
|---|
 | pmid: 26003725 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE RNA POLYMERASE I SUBCOMPLEX A14/43 |
|---|
 | pmid: 18160037 |
| 3D3D | (1) | TRANSTHYRETIN (ALSO CALLED PREALBUMIN) COMPLEX WITH THYROXINE (T4) |
|---|
 | doi: 10.1107/s0907444996003046 |
| 3D3D | (1) | TRANSTHYRETIN (ALSO CALLED PREALBUMIN) COMPLEX WITH 3',5'-DINITRO-N-ACETYL-L-THYRONINE |
|---|
 | doi: 10.1107/s0907444996003046 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF CELLULASE 12A FROM THERMOTOGA MARITIMA |
|---|
 | Q60032_THEMT | Q60032 (UniProtKB/TrEMBL) |
| 3D3D | (1) | CELLOTETRAOSE COMPLEX OF CELLULASE 12A FROM THERMOTOGA MARITIMA |
|---|
 | Q60032_THEMT | Q60032 (UniProtKB/TrEMBL) |
| 3D3D | (1) | E134C-CELLOBIOSE COMPLEX OF CELLULASE 12A FROM THERMOTOGA MARITIMA |
|---|
 | Q60032_THEMT | Q60032 (UniProtKB/TrEMBL) |
| 3D3D | (1) | E134C-CELLOTETRAOSE COMPLEX OF CELLULASE 12A FROM THERMOTOGA MARITIMA |
|---|
 | Q60032_THEMT | Q60032 (UniProtKB/TrEMBL) |
| 3D3D | (1) | E134C-CELLOBIOSE CO-CRYSTAL OF CELLULASE 12A FROM THERMOTOGA MARITIMA |
|---|
 | Q60032_THEMT | Q60032 (UniProtKB/TrEMBL) |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE FAT MASS AND OBESITY ASSOCIATED (FTO) PROTEIN REVEALS BASIS FOR ITS SUBSTRATE SPECIFICITY |
|---|
 | pmid: 20376003 |
| 3D3D | (1) | MAJOR COLD-SHOCK PROTEIN FROM ESCHERICHIA COLI SOLUTION NMR STRUCTURE |
|---|
 | expression_system_gene: u60035; gene: u60035 |
| 3D3D | (1) | Y61G MUTANT OF CELLULASE 12A FROM THERMOTOGA MARITIMA |
|---|
 | Q60032_THEMT | Q60032 (UniProtKB/TrEMBL) |
| 3D3D | (1) | Y61-GG INSERTION MUTANT OF TM-CELLULASE 12A |
|---|
 | Q60032_THEMT | Q60032 (UniProtKB/TrEMBL) |
| 3D3D | (1) | THE INSERTION MUTANT Y61GG OF TM CEL12A |
|---|
 | Q60032_THEMT | Q60032 (UniProtKB/TrEMBL) |
| 3D3D | (1) | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGMO6AATCCGCG): THE WATSON-CRICK TYPE N6-METHOXYADENOSINE/CYTIDINE BASE-PAIRS IN B-DNA |
|---|
 | pmid: 10600379 |
| 3D3D | (1) | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGMO6AATTCGCG): N6-METHOXYADENOSINE/ THYMIDINE BASE-PAIRS IN B-DNA |
|---|
 | pmid: 10600380 |
| 3D3D | (1) | STRUCTURE OF FACTOR VIIA IN COMPLEX WITH THE INHIBITOR (6S)-N-(4-CARBAMIMIDOYLBENZYL)-1-CHLORO-3-(CYCLOBUTYLAMINO)-8,8-DIETHYL-4-OXO-4,6,7,8-TETRAHYDROPYRROLO[1,2-A]PYRAZINE-6-CARBOXAMIDE |
|---|
 | pmid: 23416003 |
| 3D3D | (1) | THE STRUCTURE OF SUPERINHIBITORY PHOSPHOLAMBAN BOUND TO THE CALCIUM PUMP SERCA1A |
|---|
 | pmid: 23996003 |
| 3D3D | (1) | RACEMIC INFLUENZA M2-TM CRYSTALLIZED FROM MONOOLEIN LIPIDIC CUBIC PHASE |
|---|
 | pmid: 26460035 |
| 3D3D | (1) | RACEMIC M2-TM CRYSTALLIZED FROM RACEMIC DETERGENT |
|---|
 | pmid: 26460035 |
| 3D3D | (1) | LABORATORY EVOLVED VARIANT R-C1B1D33E6 OF POTATO EPOXIDE HYDROLASE STEH1 |
|---|
 | doi: 10.1002/cbic.201600330 |
| 3D3D | (1) | LABORATORY EVOLVED VARIANT R-C1B1D33 OF POTATO EPOXIDE HYDROLASE STEH1 |
|---|
 | doi: 10.1002/cbic.201600330 |
| 3D3D | (1) | LABORATORY EVOLVED VARIANT R-C1 OF POTATO EPOXIDE HYDROLASE STEH1 |
|---|
 | doi: 10.1002/cbic.201600330 |
| 3D3D | (1) | THE CRYOEM STRUCTURE OF HUMAN GAMMA-SECRETASE COMPLEX |
|---|
 | ref: proc.natl.acad.sci.usa v. 112 6003 2015 |
| 3D3D | (1) | AMPR EFFECTOR BINDING DOMAIN FROM CITROBACTER FREUNDII BOUND TO UDP-MURNAC-PENTAPEPTIDE |
|---|
 | organism_taxid: 1006003 |
| 3D3D | (1) | THE STRUCTURE OF PHOSPHOLAMBAN BOUND TO THE CALCIUM PUMP SERCA1A |
|---|
 | pmid: 23996003 |
| 3D3D | (1) | MNEMIOPSIS LEIDYI ML032222A IGLUR LBD GLYCINE COMPLEX |
|---|
 | pmid: 26460032 |
| 3D3D | (1) | MNEMIOPSIS LEIDYI ML032222A IGLUR LBD COMPLEX WITH ALANINE |
|---|
 | pmid: 26460032 |
| 3D3D | (1) | MNEMIOPSIS LEIDYI ML032222A IGLUR LBD D-SERINE COMPLEX |
|---|
 | pmid: 26460032 |
| 3D3D | (1) | MNEMIOPSIS LEIDYI ML032222A IGLUR LBD SERINE COMPLEX |
|---|
 | pmid: 26460032 |
| 3D3D | (1) | STRUCTURE OF UP1 BOUND TO RNA 5'-AGU-3' |
|---|
 | pmid: 26003924 |
| 3D3D | (1) | A DEHYDRATED FORM OF GLUCOSE ISOMERASE COLLECTED AT ROOM TEMPERATURE. |
|---|
 | doi: 10.1107/s2059798316003065 |
| 3D3D | (1) | A NATIVE FORM OF GLUCOSE ISOMERASE COLLECTED AT ROOM TEMPERATURE. |
|---|
 | doi: 10.1107/s2059798316003065 |
| 3D3D | (1) | A FORM OF GLUCOSE ISOMERASE COLLECTED AT 100K. |
|---|
 | doi: 10.1107/s2059798316003065 |
| 3D3D | (1) | A DEHYDRATED FORM OF GLUCOSE ISOMERASE COLLECTED AT 100K. |
|---|
 | doi: 10.1107/s2059798316003065 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF DEATH EFFECTOR DOMAIN OF CASPASE8 IN HOMO SAPIENS |
|---|
 | pmid: 26003730 |
| 3D3D | (1) | PLEUROBRACHIA BACHEI IGLUR3 LBD GLYCINE COMPLEX |
|---|
 | pmid: 26460032 |
| 3D3D | (1) | ATP SYNTHASE FROM PARACOCCUS DENITRIFICANS |
|---|
 | pmid: 26460036 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF E.COLI BRANCHING ENZYME IN COMPLEX WITH ALPHA CYCLODEXTRIN |
|---|
 | doi: 10.1107/s2059798316003272 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF ECOLI BRANCHING ENZYME WITH BETA CYCLODEXTRIN |
|---|
 | doi: 10.1107/s2059798316003272 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF ECOLI BRANCHING ENZYME WITH GAMMA CYCLODEXTRIN |
|---|
 | doi: 10.1107/s2059798316003272 |
| 3D3D | (1) | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (WT) |
|---|
 | doi: 10.1107/s2059798316003624 |
| 3D3D | (1) | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) |
|---|
 | doi: 10.1107/s2059798316003624 |
| 3D3D | (1) | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D,I211V,Y249F,S280T, Y282F,S287N,A350C,L352F) |
|---|
 | doi: 10.1107/s2059798316003624 |
| 3D3D | (1) | TGF-BETA RECEPTOR TYPE 2 KINASE DOMAIN (E431A,R433A,E485A,K488A,R493A, R495A) |
|---|
 | doi: 10.1107/s2059798316003624 |
| 3D3D | (1) | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) IN COMPLEX WITH STAUROSPORINE |
|---|
 | doi: 10.1107/s2059798316003624 |
| 3D3D | (1) | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D,I211V,Y249F,S280T, Y282F,S287N,A350C,L352F) IN COMPLEX WITH STAUROSPORINE |
|---|
 | doi: 10.1107/s2059798316003624 |
| 3D3D | (1) | TGF-BETA RECEPTOR TYPE 2 KINASE DOMAIN (E431A,R433A,E485A,K488A,R493A, R495A) IN COMPLEX WITH STAUROSPORINE |
|---|
 | doi: 10.1107/s2059798316003624 |
| 3D3D | (1) | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) IN COMPLEX WITH 3-AMINO-6-[4-(2-HYDROXYETHYL)PHENYL]-N-[4-(MORPHOLIN-4-YL)PYRIDIN-3-YL]PYRAZINE-2-CARBOXAMIDE |
|---|
 | doi: 10.1107/s2059798316003624 |
| 3D3D | (1) | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D,I211V,Y249F, S280T, Y282F,S287N,A350C,L352F) IN COMPLEX WITH 3-AMINO-6- [4-(2-HYDROXYETHYL)PHENYL]-N-[4-(MORPHOLIN-4-YL)PYRIDIN-3-YL]PYRAZINE-2-CARBOXAMIDE |
|---|
 | doi: 10.1107/s2059798316003624 |
| 3D3D | (1) | TGF-BETA RECEPTOR TYPE 2 KINASE DOMAIN (E431A,R433A,E485A,K488A,R493A, R495A) IN COMPLEX WITH 3-AMINO-6-[4-(2- HYDROXYETHYL)PHENYL]-N-[4-(MORPHOLIN-4-YL)PYRIDIN-3-YL] PYRAZINE-2-CARBOXAMIDE |
|---|
 | doi: 10.1107/s2059798316003624 |
| 3D3D | (1) | TGF-BETA RECEPTOR TYPE 2 KINASE DOMAIN (E431A,R433A,E485A,K488A,R493A, R495A) IN COMPLEX WITH AMPPNP |
|---|
 | doi: 10.1107/s2059798316003624 |
| 3D3D | (1) | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 1.31 MGY |
|---|
 | doi: 10.1107/s2059798316003351 |
| 3D3D | (1) | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 3.88 MGY |
|---|
 | doi: 10.1107/s2059798316003351 |
| 3D3D | (1) | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 6.45 MGY |
|---|
 | doi: 10.1107/s2059798316003351 |
| 3D3D | (1) | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 9.02 MGY |
|---|
 | doi: 10.1107/s2059798316003351 |
| 3D3D | (1) | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 11.6 MGY |
|---|
 | doi: 10.1107/s2059798316003351 |
| 3D3D | (1) | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 14.2 MGY |
|---|
 | doi: 10.1107/s2059798316003351 |
| 3D3D | (1) | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 16.7 MGY |
|---|
 | doi: 10.1107/s2059798316003351 |
| 3D3D | (1) | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 19.3 MGY |
|---|
 | doi: 10.1107/s2059798316003351 |
| 3D3D | (1) | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 21.9 MGY |
|---|
 | doi: 10.1107/s2059798316003351 |
| 3D3D | (1) | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 25.0 MGY |
|---|
 | doi: 10.1107/s2059798316003351 |
| 3D3D | (1) | STRUCTURE OF CALEXCITIN-GD3+ COMPLEX. |
|---|
 | doi: 10.1107/s2053230x16003526 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF PUTATIVE EXO-BETA-1,3-GALACTANASE FROM BIFIDOBACTERIUM BIFIDUM S17 |
|---|
 | doi: 10.1107/s2053230x16003617 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HEPTAPRENYL DIPHOSPHATE SYNTHASE FROM STAPHYLOCOCCUS AUREUS |
|---|
 | doi: 10.1002/cmdc.201600311 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE JM22 TCR IN COMPLEX WITH HLA-A*0201 IN COMPLEX WITH M1-F5L |
|---|
 | pmid: 27036003 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HLA-A*0201 IN COMPLEX WITH M1-F5L |
|---|
 | pmid: 27036003 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF THE JM22 TCR IN COMPLEX WITH HLA-A*0201 IN COMPLEX WITH M1-G4E |
|---|
 | pmid: 27036003 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HLA-A*0201 IN COMPLEX WITH M1-G4E |
|---|
 | pmid: 27036003 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF HLA-A*0201 IN COMPLEX WITH M1-L3W |
|---|
 | pmid: 27036003 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF LEISHMANIA MEXICANA ARGINASE IN COMPLEX WITH INHIBITOR ABHPE |
|---|
 | doi: 10.1107/s2053230x16003630 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF LEISHMANIA MEXICANA ARGINASE IN COMPLEX WITH INHIBITOR ABHDP |
|---|
 | doi: 10.1107/s2053230x16003630 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF AVIDIN COMPLEX WITH A FERROCENE BIOTIN DERIVATIVE |
|---|
 | doi: 10.1002/cplu.201600320 |
| 3D3D | (1) | CRYSTAL STRUCTURE OF 14-3-3ZETA IN COMPLEX WITH AN ALKYNE CROSS-LINKED CYCLIC PEPTIDE DERIVED FROM EXOS |
|---|
 | doi: 10.1002/cbic.201600362 |
| 3D3D | (1) | STRUCTURAL IMPACT OF SINGLE RIBONUCLEOTIDES IN DNA |
|---|
 | doi: 10.1002/cbic.201600385 |
| 3D3D | (1) | STRUCTURAL IMPACT OF SINGLE RIBONUCLEOTIDES IN DNA |
|---|
 | doi: 10.1002/cbic.201600385 |
| 3D3D | (1) | STRUCTURAL IMPACT OF SINGLE RIBONUCLEOTIDES IN DNA |
|---|
 | doi: 10.1002/cbic.201600385 |
| 3D3D | (1) | STRUCTURAL IMPACT OF SINGLE RIBONUCLEOTIDES IN DNA |
|---|
 | doi: 10.1002/cbic.201600385 |
| 3D3D | (1) | STRUCTURAL IMPACT OF SINGLE RIBONUCLEOTIDES IN DNA |
|---|
 | doi: 10.1002/cbic.201600385 |
| 3D3D | (1) | STRUCTURAL IMPACT OF SINGLE RIBONUCLEOTIDES IN DNA |
|---|
 | doi: 10.1002/cbic.201600385 |
| 3D3D | (1) | STRUCTURAL IMPACT OF SINGLE RIBONUCLEOTIDES IN DNA |
|---|
 | doi: 10.1002/cbic.201600385 |
| 3D3D | (1) | STRUCTURAL IMPACT OF SINGLE RIBONUCLEOTIDES IN DNA |
|---|
 | doi: 10.1002/cbic.201600385 |
| 3D3D | (1) | MORF DOUBLE PHD FINGER (DPF) IN COMPLEX WITH HISTONE H3K14BU |
|---|
 | pmid: 28286003 |
| 3D3D | (1) | GUANIDINE-II RIBOSWITCH P2 HAIRPIN DIMER FROM PSEUDOMONAS AERUGINOSA |
|---|
 | pmid: 28600356 |
| 3D3D | (1) | GUANIDINE-II RIBOSWITCH P2 HAIRPIN DIMER WITH 5-BROMOU SUBSTITUTION FROM PSEUDOMONAS AERUGINOSA |
|---|
 | pmid: 28600356 |
|
|