

|
|
|
The aim of this Base Pair Directory is to compile structural information on nucleic acid base pairs.
This is work in progress. We start out from the usual canonical and noncanonical base pairs with two or three hydrogen bonds and will finally include more recently discovered unusual base pairs with only one standard hydrogen bond and additional C-H...O or C-H...N contacts, water-mediated pairs, and even base pairs with no standard hydrogen bond at all. Examples for these latter pairs include:
Nucleic acids are polymers made up of repeated units, nucleotides, comprising three components:
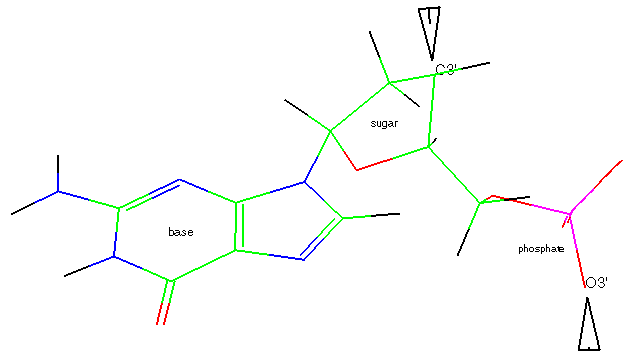
In a formal sense a nucleic acid strand is generated by forming C3'-O3'
bonds between different nucleotides. This is, however, only a formal
structural description. The chemical reaction is more complicated. The
well-known double helix is obtained by connecting the two strands via
hydrogen bonding between bases.
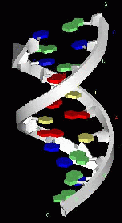
These images show a nucleic acid double helix structure in an ideal B conformation. Nucleic acids can, however, occur in different conformations. The bases are colored in the following manner: A - red, T - yellow, C - blue, G - green.
B-DNA side view
 |
B-DNA top view
 |
detailed view of a
base pair 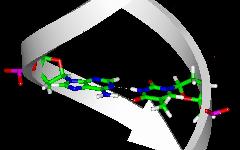 |
The bases correspond to the colored plates in the side view and are located inside in the top view.
Base pairing via hydrogen bonds as shown in the detailed view is of utmost importance for the structure of nucleic acids.
Note, however, that interactions within the sugar-phosphate backbone and base stacking are also relevant for nucleic acid structure.
The base pairs are formed from the two purine bases adenine (A) and guanine (G) and from the two pyrimidine bases cytosine (C) and uracil (U) or thymine (T).
- purine bases
|
|
| adenine - A | guanine - G |
- pyrimidine bases
|
|
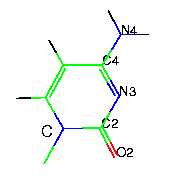 |
uracil - U |
thymine - T |
cytosine - C |
Uracil is used in RNA and thymine in DNA. The standard or canonical Watson-Crick base pairs are A-U(T) and G-C. More information on these base pairs can be found here.
In addition, other non-canonical base pairs have been found. The latter base pairs are also called mismatches. Many of them occur in RNA structures. Therefore, often only uracil but not thymine is taken into account.
There are various compilations of possible base pairs.
In 1. 28 base pairs with at least four H-bond heavy-atom donor/acceptor sites have been enumerated. The compilation 2. includes also examples with only three H-bond heavy-atom donor acceptor sites and lists 38 base pair structures. On the other hand, in 2. base pairs involving H-bonds with N3 of purines are not considered. The classification by Leontis and Westhof provides new and more comprehensive information.
In the following a comprehensive compilation is presented. The total number of possible base pairs with at least two standard H-bonds and four heavy-atom donor/acceptor site is 32. This means that four additional pairs are included as compared to the Tinoco compilation (2 x GU, 1x GG, 1 x GC). They were probably discarded for sterical reasons. However, a comprehensive search for all base pairs occurring in the currently known RNA structures has shown that this is not justified in all cases.
It is important to note that the compilations given above and below are based on simple structural rules. It cannot be excluded that a few base pairs listed do not correspond to an energy minimum. In addition, it should be kept in mind that in a nucleic acid structure stacking and backbone restraints may affect base pair geometries.
In parentheses the number of possible base pair structures with (four/three) heavy-atom donor-acceptor sites is given ( x stands for data coming soon).
purine-purine: AA (3/0) | GG (5/2) | GA (4/2) | (12/4) base pairs pyrimidine-pyrimidine: CC (2/2) | UU (3/0) | CU (2/0) | ( 7/2) base pairs purine-pyrimidine: AC (2/2) | AU (4/0) | GC (3/4) | GU (4/x) | (13/x) base pairs (not yet finalized) ---------------------------------------------------------------------------- total | (32/x) base pairs
The backbone may lead to steric restraints on base pairing. Therefore, in the preceding tables the complete nucleotides are shown. The backbone geometry corresponds to a standard A-RNA conformation. The base pair geometries were generated manually. The two bases are located approximately in a common plane and the hydrogen bond H...O or H...N distances are approximately 2 A. The structures shown do not correspond to either optimized or experimental geometries.
Both the canonical and non-canonical base pairs mentioned above were formed from standard nucleotides/bases. Modified nucleotides/bases do also occur. A few of them found in transfer RNA are shown here. A comprehensive compilation of modified nucleotides in RNA can be obtained from the RNA Modification Database.
Direct questions and criticism to Jürgen Sühnel.
|
|