

Nucleic acids are polymers made up of repeated units, nucleotides, comprising three components:
Base pairing via hydrogen bonds is of utmost importance for the structure of nucleic acids. There are, however, other relevant interactions like base stacking and interactions within the sugar-phosphate backbone. The base pairs are formed from the two purine bases adenine (A) and guanine (G) and from the two pyrimidine bases cytosine (C) and uracil (U) or thymine (T).
- purine bases
|
|
| adenine - A | guanine - G |
- pyrimidine bases
|
|
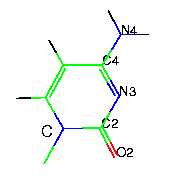 |
uracil - U |
thymine - T |
cytosine - C |
Uracil is used in RNA and thymine in DNA. In addition to the canonical Watson-Crick base pairs A-U(T) and G-C many other non-canonical base pairs have been found. The latter base pairs are also called mismatches. Many of them occur in RNA structures. Therefore, the following compilation takes only uracil but not thymine into account.
The aim of this Base Pair Directory is to compile information on nucleic acid base pairs.
In a first step tables for all possible base pairs with at least two standard hydrogen bonds are provided.
Base pairs with at least two standard hydrogen bonds
purine-purine: AA (3) | GG (5) | GA (2) | 10 base pairs
pyrimidine-pyrimidine: CC (2) | UU (3) | CU (2) | 7 base pairs
purine-pyrimidine: AC (2) | AU (4) | GC (3) | GU (4) | 15 base pairs
----------------------------------------------------------------------------
total | 32 base pairs
The backbone may lead to steric restraints on base pairing. Therefore, in the preceding tables the complete nucleotides are shown. The backbone geometry corresponds to a standard A-RNA conformation. The base pair geometries were generated manually. The two bases are located approximately in a common plane and the hydrogen bond H...O or H...N distances are approximately 2 A. The structures shown do not correspond to either optimized or experimental geometries.
Compilation of base pairs with at least two standard hydrogen bonds are also provided by:
In these cases 28 pairs are mentioned. In the preceding tables 4 additional base pairs (2 x GU, 1x GG, 1 x GC) are included. They were probably dicarded for sterical reasons. However, a comprehensive search for all base pairs occurring in the currently known RNA structures has shown that this is not justified in all cases.