

|
|
|

 1. Base Numbering
1. Base Numbering
 2. Backbone Torsion Angles in Nucleic Acid Structures
2. Backbone Torsion Angles in Nucleic Acid Structures
atomic numbering scheme and definition of
torsion angles for a polyribonucleotide chain
see also: other information on backbone torsional angles
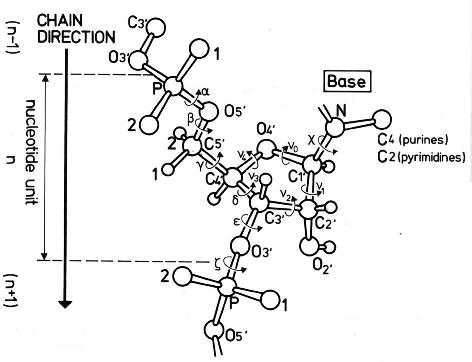
Source: Saenger,W., Principles of Nucleic Acid Structure, Springer
Verlag New York 1984.
| Average Torsion Angles for Nucleic Acid Helices (in °) | |||||||
|---|---|---|---|---|---|---|---|
| Structure Type | Alpha | Beta | Gamma | Delta | Epsilon | Zeta | Chi |
| A-DNA (fibres) | -50 | 172 | 41 | 79 | -146 | -78 | -154 |
| GGCCGGCC | -75 | 185 | 56 | 91 | -166 | -75 | -149 |
| B-DNA (fibres) | -41 | 136 | 38 | 139 | -133 | -157 | -102 |
| CGCGAATTCGCG | -63 | 171 | 54 | 123 | -169 | -108 | -117 |
| Z-DNA (C residues) | -137 | -139 | 56 | 138 | -95 | 80 | -159 |
| Z-DNA (G residues) | 47 | 179 | -169 | 99 | -104 | -69 | 68 |
| DNA-RNA decamer | -69 | 175 | 55 | 82 | -151 | -75 | -162 |
| A-RNA | -68 | 178 | 54 | 82 | -153 | -71 | -158 |

 3. Pucker
3. Pucker
 4. Chi Angle
4. Chi Angle
 5. Structures of Base Pairs Involving at Least Two Hydrogen Bonds
5. Structures of Base Pairs Involving at Least Two Hydrogen Bonds Watson-Crick, Reverse Watson-Crick, Hoogsteen, Reverse Hoogsteen, Wobble, Reverse Wobble
Watson-Crick, Reverse Watson-Crick, Hoogsteen, Reverse Hoogsteen, Wobble, Reverse Wobble Homo Purines
Homo Purines Hetero Purines
Hetero Purines Homo- and Hetero Pyrimidines
Homo- and Hetero Pyrimidines
 6. Base Triples
6. Base Triples
Proposed
hydrogen-bonding schemes for base triples.
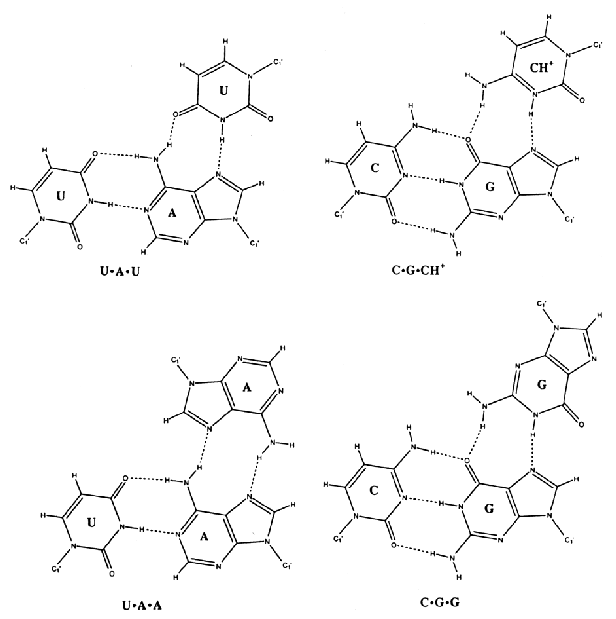
Source: Chastain, M. and Tinoco
Jr., I., (1991) Prog. Nucleic Acid Res. Mol. Biol. 41,
131-177.

 7. Movement of Bases(see also: more details on B-DNA and A-RNA)
7. Movement of Bases(see also: more details on B-DNA and A-RNA)
movements of
bases in sequence-dependent structures (tip, inclination, opening,
propeller, buckle, twist, roll, tilt, slide, rise, shift and tilt).
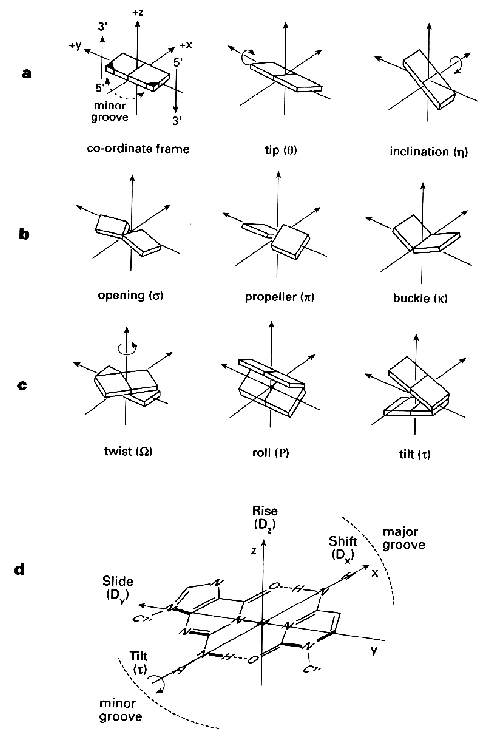
Source: Blackburn and Gait,
Nucleic acids in chemistry and biology, Oxford University Press New
York 1996.
R.E. Dickerson et al. (1989) Nucleic
Acids Res. 17, 1797-1803.

 8. Secondary Structures of RNA
8. Secondary Structures of RNA
secondary
structure of RNA consists of duplex and loop regions that can be
divided into six different types: duplexes, single-stranded regions,
hairpins, internal loops or bubbles, bulge loops or bulges and
junctions.
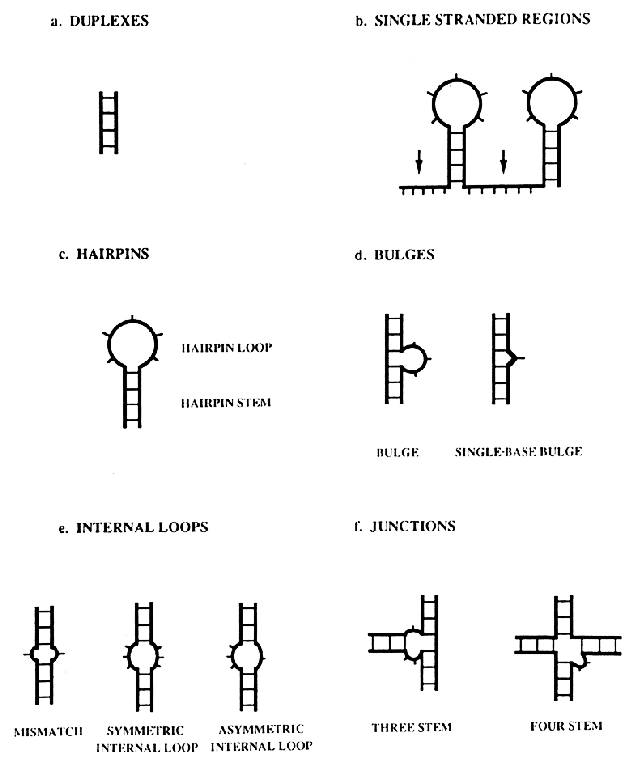
Source: Chastain, M. and Tinoco
Jr., I., (1991) Prog. Nucleic Acid Res. Mol. Biol. 41,
131-177.

 9. Pseudoknots
9. PseudoknotsRNA pseudoknots are tertiary structural elements that result when a loop in a secondary structure pairs with a complementary sequence outside the loop
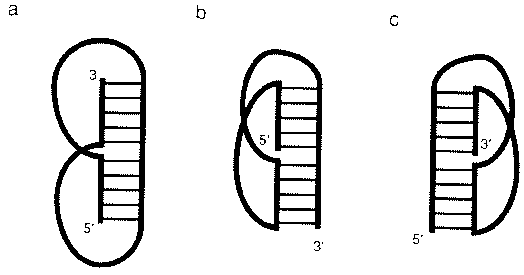
Source: Chastain, M. and Tinoco
Jr., I., (1991) Prog. Nucleic Acid Res. Mol. Biol. 41,
131-177.
The H-type pseudoknot. A pseudoknot is always defined by two stems (S1 and S2) and by two or three loop regions (L1-L3). Dashed lines indicate base-pairing. LD is the loop crossing the deep groove, LS is the loop crossing the shallow groove, and LP is the loop spanning the sugar-phosphate backbone
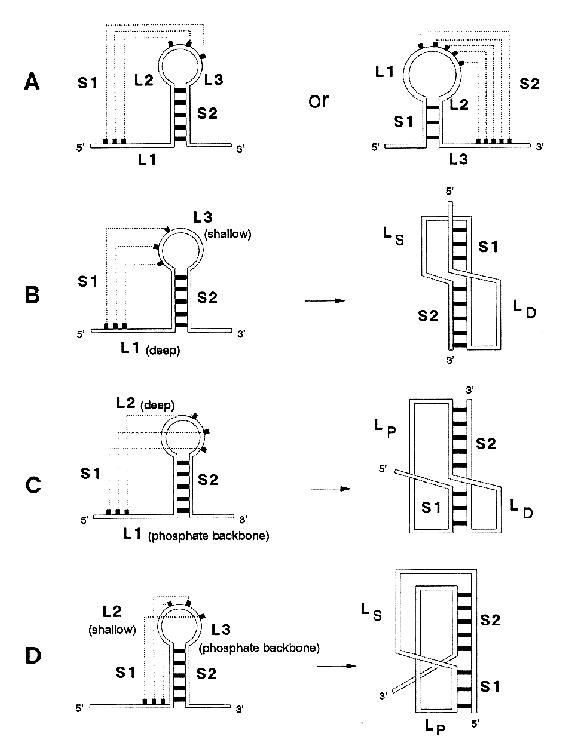
Source: Cornelis W. A. Pleij in
Gesteland, R. F. and Atkins, J. F. (1993) THE RNA WORLD. Cold Spring
Harbor Laboratory Press.
|
|