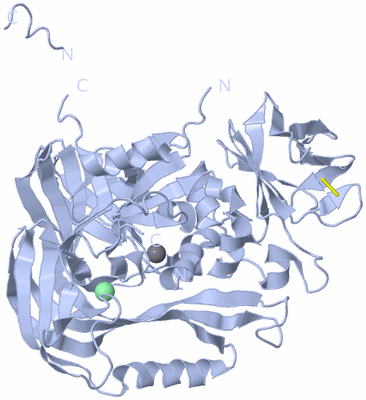
|
|
|

 Description
Description|
|

 Compounds
Compounds
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||

 Chains, Units
Chains, Units
Summary Information (see also Sequences/Alignments below) |

 Ligands, Modified Residues, Ions (2, 4)
Ligands, Modified Residues, Ions (2, 4)| Asymmetric Unit (2, 4) Biological Unit 1 (0, 0) Biological Unit 2 (0, 0) Biological Unit 3 (0, 0) |

 Sites (4, 4)
Sites (4, 4)
Asymmetric Unit (4, 4)
|

 SS Bonds (2, 2)
SS Bonds (2, 2)
Asymmetric Unit
|
||||||||||||

 Cis Peptide Bonds (3, 3)
Cis Peptide Bonds (3, 3)
Asymmetric Unit
|
||||||||||||||||

 SAPs(SNPs)/Variants (0, 0)
SAPs(SNPs)/Variants (0, 0)| (no "SAP(SNP)/Variant" information available for 3VU1) |

 PROSITE Motifs (0, 0)
PROSITE Motifs (0, 0)| (no "PROSITE Motif" information available for 3VU1) |

 Exons (0, 0)
Exons (0, 0)| (no "Exon" information available for 3VU1) |

 Sequences/Alignments
Sequences/Alignments
Asymmetric UnitChain A from PDB Type:PROTEIN Length:488 aligned with O74088_PYRHO | O74088 from UniProtKB/TrEMBL Length:976 Alignment length:496 976 496 506 516 526 536 546 556 566 576 586 596 606 616 626 636 646 656 666 676 686 696 706 716 726 736 746 756 766 776 786 796 806 816 826 836 846 856 866 876 886 896 906 916 926 936 946 956 966 976 O74088_PYRHO 487 EIEVTGWEQALKWLRSNTSKYATATSWWDYGYWIESSLLGNRRASADGGHARDRDHILALFLARDGNISEVDFESWELNYFIIYLNDWAKFNAISYLGGAITRKEYNGDENGRGRVTTILLTQAAGNVYVNPYARIVIKVIQQNKTRRIAVNIGQLECSPILSVAFPGNIKIKGSGRCSDGSPFPYVVYLTPSLGVLAYYKVATSNFVKLAFGIPTSSYSEFAEKLFSNFIPVYQYGSVIVYEFRPFAIYKIEDFINGTWREVGKLSPGKHTLRLYISAFGRDIKNATLYVYALNGTKIIKRIKVGEIKYMNHLEEYPIIVNVTLPTAQKYRFILAQKGPVGVLTGPVRVNGKITNPAYIMREGESGRLELKVGVDKEYTADLYLRATFIYLVRKGGKSNEDYDASFEPHMDTFFITKLKEGIKLRPGENEIVVNAEMPKNAISSYKEKLEKEHGDKLIIRGIRVEPVFIVEKEYTMIEVSASAPHHSSE------ - SCOP domains ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- SCOP domains CATH domains ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CATH domains Pfam domains ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- Pfam domains Sec.struct. author .....hhhhhhhhhhhhh.....eee.................--------.hhhhhhhhhhhh.hhhhh.hhhhhh..eeeee.hhhhhhhhhhhhh...hhhhhhh..........eeeeee...eeee....eeeeeee....eeeeeee..eeee..eeee....eeee...ee.......eeeee...eeeeee.hhh.hhhhhhhhh.......hhhhhhhhheeeeeee..eeeeee..eeeeeeeeee..eeee......eeeeeeeeeee....eeeeeeeeeeee..eeeeeeeeeeeeee.......eeeeeee....eeeeeeeeeeee.ee...eee..eee..........eeeeeeeeee...eeeeeeeeeeeeeeeee..........eeeeeeeeeeeeeeeeeeee..eeeeeeeeee...hhhhhhhhhhhhhhh..eeeeeeeeeeeeeeeeeeeeeeee............... Sec.struct. author SAPs(SNPs) ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- SAPs(SNPs) PROSITE ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- PROSITE Transcript ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- Transcript 3vu1 A 487 EIEVTGWEQALKWLRSNTSKYATATSWWDYGYWIESSLLGNRR--------RDRDHILALFLARDGNISEVDFESWELNYFIIYLNDWAKFNAISYLGGAITRKEYNGDENGRGRVTTILLTQAAGNVYVNPYARIVIKVIQQNKTRRIAVNIGQLECSPILSVAFPGNIKIKGSGRCSDGSPFPYVVYLTPSLGVLAYYKVATSNFVKLAFGIPTSSYSEFAEKLFSNFIPVYQYGSVIVYEFRPFAIYKIEDFINGTWREVGKLSPGKHTLRLYISAFGRDIKNATLYVYALNGTKIIKRIKVGEIKYMNHLEEYPIIVNVTLPTAQKYRFILAQKGPVGVLTGPVRVNGKITNPAYIMREGESGRLELKVGVDKEYTADLYLRATFIYLVRKGGKSNEDYDASFEPHMDTFFITKLKEGIKLRPGENEIVVNAEMPKNAISSYKEKLEKEHGDKLIIRGIRVEPVFIVEKEYTMIEVSASAPHHHHHHHHHHH 986 496 506 516 526 | - | 546 556 566 576 586 596 606 616 626 636 646 656 666 676 686 696 706 716 726 736 746 756 766 776 786 796 806 816 826 836 846 856 866 876 886 896 906 916 926 936 946 956 966 |980 529 538 973| 978 Chain B from PDB Type:PROTEIN Length:490 aligned with O74088_PYRHO | O74088 from UniProtKB/TrEMBL Length:976 Alignment length:490 495 505 515 525 535 545 555 565 575 585 595 605 615 625 635 645 655 665 675 685 695 705 715 725 735 745 755 765 775 785 795 805 815 825 835 845 855 865 875 885 895 905 915 925 935 945 955 965 975 O74088_PYRHO 486 TEIEVTGWEQALKWLRSNTSKYATATSWWDYGYWIESSLLGNRRASADGGHARDRDHILALFLARDGNISEVDFESWELNYFIIYLNDWAKFNAISYLGGAITRKEYNGDENGRGRVTTILLTQAAGNVYVNPYARIVIKVIQQNKTRRIAVNIGQLECSPILSVAFPGNIKIKGSGRCSDGSPFPYVVYLTPSLGVLAYYKVATSNFVKLAFGIPTSSYSEFAEKLFSNFIPVYQYGSVIVYEFRPFAIYKIEDFINGTWREVGKLSPGKHTLRLYISAFGRDIKNATLYVYALNGTKIIKRIKVGEIKYMNHLEEYPIIVNVTLPTAQKYRFILAQKGPVGVLTGPVRVNGKITNPAYIMREGESGRLELKVGVDKEYTADLYLRATFIYLVRKGGKSNEDYDASFEPHMDTFFITKLKEGIKLRPGENEIVVNAEMPKNAISSYKEKLEKEHGDKLIIRGIRVEPVFIVEKEYTMIEVSASAPHHSS 975 SCOP domains ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- SCOP domains CATH domains ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CATH domains Pfam domains ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- Pfam domains Sec.struct. author ......hhhhhhhhhhhhh.....eee.........................hhhhhhhhhhhhh.hhhhh.hhhhhh..eeeee.hhhhhhhhhhhhh...hhhhhhh..........eeeeeee..eeee....eeeeeee....eeeeeee..eeee..eeee....eeee...ee.......eeeee...eeeeee.hhh.hhhhhhhhh.......hhhhhhhhheeeeeee..eeeeee..eeeeeeeeee..eeee......eeeeeeeeeee....eeeeeeeeeeee..eeeeeeeeeeeeee.......eeeeeee....eeeeeeeeeeee.eeeeeeee..eee..........eeeeeeeeee.....eeeeeeeeeeeeeee..........eeeeeeeeeeeeeeeeee....eeeeeeeeee....hhhhhhhhhhhhhh..eeeeeeeeeeeeeeeeeeeeeeee........ Sec.struct. author SAPs(SNPs) ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- SAPs(SNPs) PROSITE ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- PROSITE Transcript ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- Transcript 3vu1 B 486 TEIEVTGWEQALKWLRSNTSKYATATSWWDYGYWIESSLLGNRRASADGGHARDRDHILALFLARDGNISEVDFESWELNYFIIYLNDWAKFNAISYLGGAITRKEYNGDENGRGRVTTILLTQAAGNVYVNPYARIVIKVIQQNKTRRIAVNIGQLECSPILSVAFPGNIKIKGSGRCSDGSPFPYVVYLTPSLGVLAYYKVATSNFVKLAFGIPTSSYSEFAEKLFSNFIPVYQYGSVIVYEFRPFAIYKIEDFINGTWREVGKLSPGKHTLRLYISAFGRDIKNATLYVYALNGTKIIKRIKVGEIKYMNHLEEYPIIVNVTLPTAQKYRFILAQKGPVGVLTGPVRVNGKITNPAYIMREGESGRLELKVGVDKEYTADLYLRATFIYLVRKGGKSNEDYDASFEPHMDTFFITKLKEGIKLRPGENEIVVNAEMPKNAISSYKEKLEKEHGDKLIIRGIRVEPVFIVEKEYTMIEVSASAPHHSS 975 495 505 515 525 535 545 555 565 575 585 595 605 615 625 635 645 655 665 675 685 695 705 715 725 735 745 755 765 775 785 795 805 815 825 835 845 855 865 875 885 895 905 915 925 935 945 955 965 975
|
||||||||||||||||||||

 SCOP Domains (0, 0)
SCOP Domains (0, 0)| (no "SCOP Domain" information available for 3VU1) |

 CATH Domains (0, 0)
CATH Domains (0, 0)| (no "CATH Domain" information available for 3VU1) |

 Pfam Domains (0, 0)
Pfam Domains (0, 0)| (no "Pfam Domain" information available for 3VU1) |

 Gene Ontology (5, 5)
Gene Ontology (5, 5)|
Asymmetric Unit(show GO term definitions) Chain A,B (O74088_PYRHO | O74088)
|
||||||||||||||||||||||||||||||||||||||||||||||||

 Interactive Views
Interactive Views
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||

 Still Images
Still Images
|
||||||||||||||||

 Databases
Databases
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||

 Analysis Tools
Analysis Tools
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||

 Entries Sharing at Least One Protein Chain (UniProt ID)
Entries Sharing at Least One Protein Chain (UniProt ID)| (no "Entries Sharing at Least One Protein Chain" available for 3VU1) |

 Related Entries Specified in the PDB File
Related Entries Specified in the PDB File
|
|