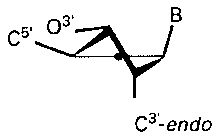Helix and bending analysis of nucleic acid double helix structures
Nucleic acid backbone parameters
1. Backbone torsion angles
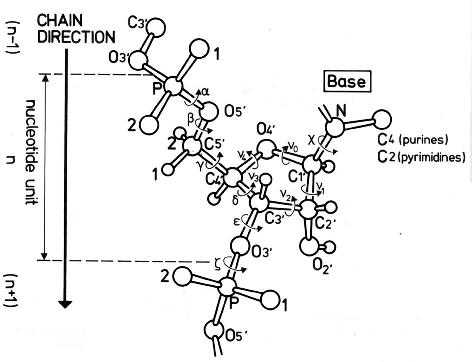 Figure 1. Backbone torsion angles in nucleic acid
structures. (Saenger W. 1984. Principles of Nucleic Acid Structure.
Springer, New York.)
Figure 1. Backbone torsion angles in nucleic acid
structures. (Saenger W. 1984. Principles of Nucleic Acid Structure.
Springer, New York.)
The torsion angles in the backbone of nucleic acid chains are designated with
greek letters (alpha to zeta). Because the torsion
angles are correlated, we list only a subset of the torsion angles in
the table. The angles selected are most informative for describing the
conformation of double helices.
alpha, beta
Alpha is the torsion angle about the P-O5' bond, and
beta is about the O5'-C5' bond. Beta is found
almost always in the antiperiplanar range
(+150° ... +210°).
gamma
Gamma is the torsion angle about the C5'-C4' bond.
The +synclinal range (+30° ... +90°) is preferred
in most duplex structures. In Z-DNA however, syn guanines
show gamma in the antiperiplanar range
(+150° ... +210°).
delta
Delta is the torsion angle about the C4'-C3' bond.
Delta is correlated with the sugar pucker phase angle
(see 2. Sugar pucker).
epsilon-zeta (BI/BII conformation of phosphate)
Epsilon is the torsion angle about the C3'-O3' bond, and
zeta is about the O3'-P bond. Both angles determine the position
of the phosphate with respect to the sugar of the preceding residue.
A phosphate can adopt either the BI or the BII conformation.
The BI conformation is most often found in double helices.
Nevertheless, some phosphate groups swap temporarily into
the BII conformation. These residues show a dynamic equilibrium between
BI and BII conformation. Sometimes such residues are caught in
the BII conformation in time averaged structures as obtained
from X-ray crystallography or NMR.
The difference (epsilon-zeta) is commonly used to
characterize the conformation of the phosphate group. Here we use
the following assignment:
| BI: |
(epsilon-zeta) |
= |
-160° ... +20° |
| BII: |
(epsilon-zeta) |
= |
+20° ... +200° |
A phosphate in the BII conformation
forces its neighboring phosphates into the BI conformation.
Having two neighboring phosphates in the BII conformation at the same
time is an extremely rare event.
2. Sugar pucker
Figure 2. Structures for the C2'-endo and C3'-endo conformations
of ribose. (Blackburn GM, Gait MJ. 1996. Nucleic acids in chemistry and biology.
Oxford University Press.)
The conformation of the ribose ring can be described by only two
quantities, sugar pucker phase angle and sugar pucker amplitude.
Pucker phase angle and pucker amplitude are calculated from the torsion
angles about the five bonds in the ribose ring. The torsion angles in
a closed ring are highly correlated.
The differing conformations of the ribose have been named
with respect to that ring atom which puckers out of the plane given by
the other ring atoms. The most prominent conformations are C2'-endo in
B-helices and C3'-endo in A-helices. At room temperature both
conformers are in a dynamic equilibrium. Transitions occur via the
O4'-endo state. Intermediates between C2'-endo and C3'-endo are found
in several (time-averaged) structures obtained from X-ray
crystallography or NMR. When changing the pucker phase angle from
0° to 360° we step through all possible conformations of the
ribose ring. This pseudo-rotation cycle can be depicted as a
conformational wheel (Figure 3).
Table 1. Ranges of the sugar pucker phase angle
for all conformations of ribose.
| Angle range |
Name |
Occurrence |
|---|
|
|
|
C2'-exo |
|
|
|
|
C3'-endo |
ideal A-helix |
|
|
|
C4'-exo |
|
|
|
|
O4'-endo |
|
|
|
|
C1'-exo |
|
|
|
|
C2'-endo |
ideal B-helix |
|
|
|
C3'-exo |
|
|
|
|
C4'-endo |
|
|
|
|
O4'-exo |
|
|
|
|
C1'-endo |
|
|
|
|
C2'-exo |
|
 Figure 3. Conformational wheel of the sugar pucker pseudo-rotation cycle
(Saenger W. 1984. Principles of Nucleic Acid Structure. Springer, New York.)
Figure 3. Conformational wheel of the sugar pucker pseudo-rotation cycle
(Saenger W. 1984. Principles of Nucleic Acid Structure. Springer, New York.)
3. Glycosidic bond
chi (syn and anti conformation)
Chi is the torsion angle about the N-glycosidic bond between the
ribose and purine or pyrimidine base. Rotamers with chi values
between -90° and +90° are called syn whereas anti
refers to chi values from +90° to +270°. In
Watson-Crick double helixes both bases of a base pairs are in
anti conformation. Bases in syn conformation indicate a
distortion of the double helix due to base pair opening or mismatched
base pairs. The syn conformation is also found with guanines in
left-handed Z-DNA (zig-zag helix). Calculation of the helical axis with
Curves 5.3 fails for technical reasons, if a right-handed double
helix contains bases in syn conformation.
 Figure 4. Torsional energy profile for rotation about the
glycosidic bond in desoxyribocytosine. Energies were calculated with a
molecular mechanics force field (Cornell et al. 1995. J Am Chem Soc, 117: 5179-5197.)
applying vacuum conditions.
Figure 4. Torsional energy profile for rotation about the
glycosidic bond in desoxyribocytosine. Energies were calculated with a
molecular mechanics force field (Cornell et al. 1995. J Am Chem Soc, 117: 5179-5197.)
applying vacuum conditions.
The typical energy profile for rotation about the glycosidic bond of
a desoxyribonucleotide in Figure 4. It turns out that the
energy minimum of the anti conformation is located at -140°
and the minimum of the syn conformation at +30°. In
addition, there is a third minimum at -60°. The range between
-110° and -60° is termed high anti. The high
anti range is often populated in duplex structures.
The glycosidic torsion angle chi is correlated with sugar
puckering. High anti is often found with C2'-endo sugars
(B-helix), while C3'-endo sugars (A-helix) are associated with more
canonical anti values in the range from -150° to -180°.
Top of page Close window